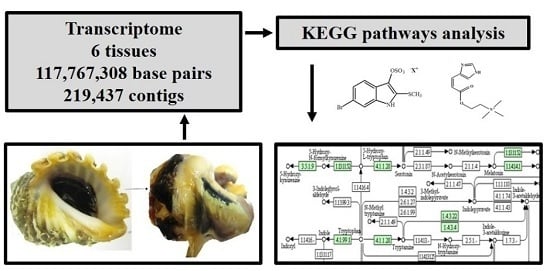
Transcriptome of the Australian Mollusc Dicathais orbita Provides Insights into the Biosynthesis of Indoles and Choline Esters
Abstract
:1. Introduction
2. Results and Discussion
2.1. De Novo Transcriptome Assembly
2.2. Transcriptome Annotation
2.3. Tryptophan Metabolism and Phenylalanine, Tyrosine, Tryptophan Biosynthetic Pathways
2.4. Sulfur, Cysteine and Methionine Metabolisms Pathway in Dicathais orbita
2.5. Bromoperoxidase Enzymes
2.6. Dicathais Orbita Glycerophospholipid and Histidine Metabolism Pathway
3. Materials and Methods
3.1. Specimen Collection
3.2. Transcriptome Sequencing
3.3. De Novo Transcriptome Assembly and Annotation
3.4. Nucleotide Sequence Accession Number
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| ENA | European Nucleotide Archive |
| NCBI | National Center for Biotechnology Information |
| BLAST | Basic Local Alignment Search Tool |
| ORFs | Open Reading Frames |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
References
- Westley, C.; Benkendorff, K. Sex-specific Tyrian purple genesis: Precursor and pigment distribution in the reproductive system of the marine mollusc, Dicathais orbita. J. Chem. Ecol. 2008, 34, 44–56. [Google Scholar] [CrossRef] [PubMed]
- Benkendorff, K.; Rudd, D.; Nongmaithem, B.D.; Liu, L.; Young, F.; Edwards, V.; Avila, C.; Abbott, C.A. Are the traditional medical uses of Muricidae molluscs substantiated by their pharmacological properties and bioactive compounds? Mar. Drugs 2015, 13, 5237–5275. [Google Scholar] [CrossRef] [PubMed]
- Benkendorff, K. Natural product research in the Australian marine invertebrate Dicathais orbita. Mar. Drugs 2013, 11, 1370–1398. [Google Scholar] [CrossRef] [PubMed]
- Baker, J.T.; Duke, C.C. Isolation of choline and choline ester salts of tyrindoxyl sulphate from the marine molluscs Dicathais orbita and Mancinella keineri. Tetrahedron Lett. 1976, 15, 1233–1234. [Google Scholar] [CrossRef]
- Esmaeelian, B.; Abbott, C.A.; le Leu, R.K.; Benkendorff, K. 6-bromoisatin found in muricid mollusc extracts inhibits colon cancer cell proliferation and induces apoptosis, preventing early stage tumor formation in a colorectal cancer rodent model. Mar. Drugs 2014, 12, 17–35. [Google Scholar] [CrossRef] [PubMed]
- Benkendorff, K.; Bremner, J.B.; Davis, A.R. Tyrian purple precursors in the egg masses of the Australian muricid, Dicathais orbita: A possible defensive role. J. Chem. Ecol. 2000, 26, 1037–1050. [Google Scholar] [CrossRef]
- Esmaeelian, B.; Benkendorff, K.; Johnston, M.R.; Abbott, C.A. Purified brominated indole derivatives from Dicathais orbita induce apoptosis and cell cycle arrest in colorectal cancer cell lines. Mar. Drugs 2013, 11, 3802–3822. [Google Scholar] [CrossRef] [PubMed]
- Edwards, V.; Benkendorff, K.; Young, F. Marine compounds selectively induce apoptosis in female reproductive cancer cells but not in primary-derived human reproductive granulosa cells. Mar. Drugs 2012, 10, 64–83. [Google Scholar] [CrossRef] [PubMed]
- Westley, C.B.; McIver, C.M.; Abbott, C.A.; le Leu, R.K.; Benkendorff, K. Enhanced acute apoptotic response to azoxymethane-induced DNA damage in the rodent colonic epithelium by Tyrian purple precursors: A potential colorectal cancer chemopreventative. Cancer Biol. Ther. 2010, 9, 371–379. [Google Scholar] [CrossRef] [PubMed]
- Freidlander, P. Ueber den farbstoff des antiken purpura aus Murex brandaris. Chem. Ber. 1909, 42, 765–770. [Google Scholar] [CrossRef]
- Zhang, W.; Li, F.; Nie, L. Integrating multiple “omics” analysis for microbial biology: Application and methodologies. Microbiology 2010, 156, 287–301. [Google Scholar] [CrossRef] [PubMed]
- Brown, T.A. Chapter 3: Genomes. In Transcriptomes and Proteomes, 2nd ed.; John Wiley & Sons: New York, NY, USA, 2002; pp. 69–91. [Google Scholar]
- Adams, J. Transcriptome: Connecting the genome to gene function. Nat. Educ. 2008, 1, 195. Available online: http://www.nature.com/scitable/topicpage/transcriptome-connecting-the-genome-to-gene-function-605 (accessed on 14 May 2016). [Google Scholar]
- Jackson, D.J.; McDougall, C.; Green, K.; Simpson, F.; Worheide, G.; Degnan, B.M. A rapidly evolving secretome builds and patterns a sea shell. BMC Biol. 2006, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lockyer, A.E.; Spinks, J.; Kane, R.A.; Hoffmann, K.F.; Fitzpatrick, J.M.; Rollinson, D.; Noble, L.R.; Jones, C.S. Biomphalaria glabrata transcriptome: cDNA microarray profiling identifies resistant- and susceptible-specific gene expression in haemocytes from snail strains exposed to Schistosoma mansoni. BMC Genom. 2008, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knight, M.; Arican-Goktas, H.D.; Ittiprasert, W.; Odoemelam, E.C.; Miller, A.N.; Bridger, J.M. Schistosomes and snails: A molecular encounter. Front. Genet. 2014, 5, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Moroz, L.L.; Edwards, J.R.; Puthanveettil, S.V.; Kohn, A.B.; Ha, T.; Heyland, A.; Knudsen, B.; Sahni, A.; Yu, F.; Liu, L.; et al. Neuronal Transcriptome of Aplysia: Neuronal Compartments and Circuitry. Cell 2006, 127, 1453–1467. [Google Scholar] [CrossRef] [PubMed]
- Sadamoto, H.; Takahashi, H.; Okada, T.; Kenmoku, H.; Toyota, M.; Asakawa, Y. De novo sequencing and transcriptome analysis of the central nervous system of mollusc Lymnaea stagnalis by deep RNA sequencing. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.P.; Zhang, Z.; van Kesteren, R.E.; Straub, V.A.; van Nierop, P.; Jin, K.; Nejatbakhsh, N.; Goldberg, J.I.; Spencer, G.E.; Yeoman, M.S.; et al. Transcriptome analysis of the central nervous system of the mollusc Lymnaea stagnalis. BMC Genom. 2009, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romero, A.; Novoa, B.; Figueras, A. Genomics, immune studies and diseases in bivalve aquaculture. J. Invertebr. Pathol. 2012, 9, 110–121. [Google Scholar]
- Chavez, M.J.; Valenzuela, M.V.; Nunez, A.G.; Maldonado, A.W.; Gallardo, E.C. Concholepas concholepas Ferritin H-like subunit (CcFer): Molecular characterization and single nucleotide polymorphism associated to innate immune response. Fish Shellfish Immunol. 2013, 35, 910–917. [Google Scholar] [CrossRef] [PubMed]
- Powell, D.; Knibb, W.; Remilton, C.; Elizur, A. De-novo transcriptome analysis of the banana shrimp (Fenneropenaeus merguiensis) and identification of genes associated with reproduction and development. Mar. Genom. 2015, 22, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Ho, K.K.Y.; Leung, P.T.Y.; Ip, J.C.H.; Qiu, J.W.; Leung, K.M.Y. De novo transcriptomic profile in the gonadal tissues of the intertidal whelk Reishia clavigera. Mar. Pollut. Bull. 2014, 85, 499–504. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Fang, X.; Guo, X.; Li, L.; Luo, R.; Xu, F.; Yang, P.; Zhang, L.; Wang, X.; Qi, H.; et al. The oyster genome reveals stress adaptation and complexity of shell formation. Nature 2012, 490, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Meng, J.; Zhu, Q.; Zhang, L.; Li, C.; Li, L.; She, Z.; Huang, B.; Zhang, G. Genome and transcriptome analyses provide insight into the euryhaline adaptation mechanism of Crassostrea gigas. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Cardenas, L.; Sanchez, R.; Gomez, D.; Fuenzalida, G.; Gallardo-Escarate, C.; Tanguy, A. Transcriptome analysis in Concholepas concholepas (Gastropoda, Muricidae): Mining and characterization of new genomic and molecular markers. Mar. Genom. 2011, 4, 197–205. [Google Scholar] [CrossRef] [PubMed]
- Modica, M.V.; Lombardo, F.; Franchini, P.; Oliverio, M. The venomous cocktail of the vampire snail Colubraria reticulata (Mollusca, Gastropoda). BMC Genom. 2015, 16, 441. [Google Scholar] [CrossRef] [PubMed]
- Terrat, Y.; Biass, D.; Dutertre, S.; Favreau, P.; Remm, M.; Stocklin, R.; Piquemal, D.; Ducancel, F. High-resolution picture of a venom gland transcriptome: Case study with the marine snail Conus consors. Toxicon 2012, 59, 34–46. [Google Scholar] [CrossRef] [PubMed]
- Barghi, N.; Concepcion, G.P.; Olivera, B.M.; Lluisma, A.O. Comparison of the venom peptides and their expression in closely related Conus species: Insights into adaptive post-speciation evolution of Conus exogenomes. Genome Biol. Evol. 2015, 7, 1797–1814. [Google Scholar] [CrossRef] [PubMed]
- Watkins, M.; Hillyard, D.R.; Olivera, B.M. Genes expressed in a turrid venom duct: Divergence and similarity to conotoxins. J. Mol. Evol. 2006, 62, 247–256. [Google Scholar] [CrossRef] [PubMed]
- Urbarova, I.; Karlsen, B.O.; Okkenhaug, S.; Seternes, O.M.; Johansen, S.D.; Emblem, A. Digital marine bioprospecting: Mining new neurotoxin drug candidates from the transcriptomes of cold-water sea anemones. Mar. Drugs 2012, 10, 2265–2279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Zhang, S.F.; Lin, L.; Wang, D.Z. Comparative transcriptome analysis of a toxin-producing dinoflagellate Alexandrium catenella and its non-toxic mutant. Mar. Drugs 2014, 12, 5698–5718. [Google Scholar] [CrossRef] [PubMed]
- Laffy, P.W.; Benkendorff, K.; Abbott, C.A. Suppressive subtractive hybridisation transcriptomics provides a novel insight into the functional role of the hypobranchial gland in a marine mollusc. Comp. Biochem. Physiol. D Genom. Proteom. 2013, 8, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A quality control tool for high throughput sequence data. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 14 May 2016).
- Prentis, P.J.; Pavasovic, A. The Anadara trapezia transcriptome: A resource for molluscan physiological genomics. Mar. Genom. 2014, 18, 113–115. [Google Scholar] [CrossRef] [PubMed]
- Ngangbam, A.K.; Baten, A.; Waters, D.L.E.; Whalan, S.; Benkendorff, K. Characterization of bacterial communities associated with the Tyrian purple producing gland in a marine gastropod. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- Ngangbam, A.K.; Waters, D.L.E.; Whalan, S.; Baten, A.; Benkendorff, K. Indole producing bacteria from the biosynthetic organs of Muricid mollusc could contribute to Tyrian purple production. J. Shellfish Res. 2015, 34, 443–454. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic. Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Banoglu, E.; Jha, G.G.; King, R.S. Hepatic microsomal metabolism of indole to indoxyl, a precursor of indoxyl sulfate. Eur. J. Drug. Metab. Pharmacokinet. 2001, 26, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Arora, P.K.; Sharma, A.; Bae, H. Microbial degradation of indole and its derivatives. J. Chem. 2015, 2015, 1–13. [Google Scholar] [CrossRef]
- O’Connor, K.E.; Hartmans, S. Indigo formation by aromatic hydrocarbon-degrading bacteria. Biotechnol. Lett. 1998, 20, 219–223. [Google Scholar] [CrossRef]
- Han, X.; Wang, W.; Xiao, X. Microbial biosynthesis and biotransformation of indigo and indigo-like pigments. Chin. J. Biotechnol. 2008, 24, 921–926. [Google Scholar] [CrossRef]
- Westley, C.; Vine, K.; Benkendorff, K. A proposed functional role for indole derivatives in reproduction and defense of the Muricidae (Neogastropoda: Mollusca). In Indirubin, the Red Shade of Indigo; Meijer, L., Guyard, N., Skaltsounis, L., Eisenbrand, G., Eds.; Life in Progress: Roscoff, France, 2006; pp. 31–44. [Google Scholar]
- Verhecken, A. The indole pigments of Mollusca. Ann. Soc. R. Zool. Belg. 1989, 119, 181–197. [Google Scholar]
- Westley, C.; Benkendorff, K. The distribution of precursors and biosynthetic enzymes required for Tyrian purple genesis in the hypobranchial gland, gonoduct, an egg masses of Dicathais orbita (Gmelin, 1791) (Neogastropoda: Muricidae). Nautilus 2009, 123, 148–153. [Google Scholar]
- Jannun, R.; Coe, E.L. Bromoperoxidase from the marine snail, Murex-trunculus. Comp. Biochem. Physiol. B Comp. Biochem. 1987, 88, 917–922. [Google Scholar] [CrossRef]
- Quastel, J.H.; Tennenbaum, M.; Wheatley, A.H. Choline ester formation in, and choline esterase activities of, tissues in vitro. Biochem. J. 1936, 30, 1668–1681. [Google Scholar] [CrossRef] [PubMed]
- Roseghini, M.; Severini, C.; Erspamer, G.F.; Erspamer, V. Choline esters and biogenic amines in the hypobranchial gland of 55 molluscan species of the neogastropod Muricoidea Superfamily. Toxicon 1996, 34, 33–55. [Google Scholar] [CrossRef]
- Sundberg, R.J.; Martin, R.B. Interactions of histidine and other imidazole derivatives with transition metal ions in chemical and biological systems. Chem. Rev. 1974, 74, 471–517. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef] [PubMed]
- Zdobnov, E.M.; Apweiler, R. InterProScan—An integration platform for the signature-recognition methods in InterPro. Bioinformatics 2001, 17, 847–848. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Fang, L.; Zheng, H.; Zhang, Y.; Chen, J.; Zhang, Z.; Wang, J.; Li, S.; Li, R.; Bolund, L.; et al. WEGO: A web tool for plotting GO annotations. Nucleic Acids Res. 2006, 34, W293–W297. [Google Scholar] [CrossRef] [PubMed]









| Snails (S) | Description | Raw Reads | High Quality Reads | |
|---|---|---|---|---|
| Number | Percent | |||
| S1 + S2 + S3 | Female hypobranchial gland 1, August, Breeding season, 2014 | 15,531,322 | 15,100,466 | 97.23 |
| S4 + S5 + S6 | Female hypobranchial gland 2, August, Breeding season, 2014 | 15,693,385 | 15,258,671 | 97.23 |
| S7 + S8 + S9 | Female hypobranchial gland 1, January, 2015 | 15,835,271 | 15,425,533 | 97.41 |
| S10 + S11 + S12 | Female hypobranchial gland 2, January, 2015 | 16,457,635 | 15,990,724 | 97.16 |
| S13 + S14 + S15 | Male hypobranchial gland 1, January, 2015 | 16,142,317 | 15,684,926 | 97.17 |
| S16 + S17 + S18 | Male hypobranchial gland 2, January, 2015 | 17,461,007 | 16,997,497 | 97.35 |
| S7 + S8 + S9 | Female foot 1, January, 2015 | 16,015,535 | 15,595,463 | 97.38 |
| S10 + S11 + S12 | Female foot 2, January, 2015 | 17,057,433 | 16,653,222 | 91.40 |
| S13 + S14 + S15 | Male foot 1, January, 2015 | 14,241,690 | 13,885,327 | 97.50 |
| S16 + S17 + S18 | Male foot 2, January, 2015 | 15,813,363 | 15,406,030 | 97.42 |
| S7 + S8 + S9 | Capsule gland, January, 2015 | 15,805,867 | 15,291,498 | 96.75 |
| S7 + S8 + S9 | Albumen gland, January, 2015 | 14,442,864 | 14,011,099 | 97.01 |
| S13 + S14 + S15 | Prostate gland, January, 2015 | 15,600,688 | 15,113,842 | 96.88 |
| S10 + S11 + S12 | Mantle 1, January, 2015 | 16,273,556 | 15,804,247 | 97.12 |
| - | Total | 222,371,933 | 216,218,545 | - |
| Contig Summary Statistics | bp (Base Pair) |
|---|---|
| Number of contigs | 219,437 |
| Total assembly length | 117,767,308 |
| N50 | 608 |
| Mean contig length | 537 |
| Largest contig length | 12,897 |
| Number of contigs larger than 500 bp | 59,144 |
| Number of contigs larger than 1000 bp | 22,818 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baten, A.; Ngangbam, A.K.; Waters, D.L.E.; Benkendorff, K. Transcriptome of the Australian Mollusc Dicathais orbita Provides Insights into the Biosynthesis of Indoles and Choline Esters. Mar. Drugs 2016, 14, 135. https://doi.org/10.3390/md14070135
Baten A, Ngangbam AK, Waters DLE, Benkendorff K. Transcriptome of the Australian Mollusc Dicathais orbita Provides Insights into the Biosynthesis of Indoles and Choline Esters. Marine Drugs. 2016; 14(7):135. https://doi.org/10.3390/md14070135
Chicago/Turabian StyleBaten, Abdul, Ajit Kumar Ngangbam, Daniel L. E. Waters, and Kirsten Benkendorff. 2016. "Transcriptome of the Australian Mollusc Dicathais orbita Provides Insights into the Biosynthesis of Indoles and Choline Esters" Marine Drugs 14, no. 7: 135. https://doi.org/10.3390/md14070135
APA StyleBaten, A., Ngangbam, A. K., Waters, D. L. E., & Benkendorff, K. (2016). Transcriptome of the Australian Mollusc Dicathais orbita Provides Insights into the Biosynthesis of Indoles and Choline Esters. Marine Drugs, 14(7), 135. https://doi.org/10.3390/md14070135