Blue-Print Autophagy: Potential for Cancer Treatment
Abstract
:1. Introduction to Autophagy
2. Autophagy and Cancer
3. Autophagy and Marine Natural Products
3.1. Autophagy-Inducers Marine Natural Products
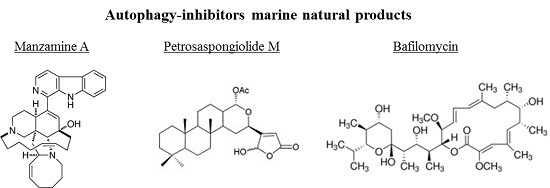
3.2. Autophagy-Inhibitors Marine Natural Products
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Alam, J.J. Apoptosis: Target for novel drugs. Trends Biotechnol. 2003, 21, 479–483. [Google Scholar] [CrossRef] [PubMed]
- Reed, J.C. Apoptosis-targeted therapies for cancer. Cancer Cell 2003, 3, 17–22. [Google Scholar] [CrossRef]
- Lockshin, R.A.; Zakeri, Z. Apoptosis, autophagy, and more. Int. J. Cell. Biol. 2004, 36, 2405–2419. [Google Scholar] [CrossRef] [PubMed]
- Trisciuoglio, D.; Uranchimeg, B.; Cardellina, J.H.; Meragelman, T.L.; Matsunaga, S.; Fusetani, N.; Del Bufalo, D.; Shoemaker, R.H.; Melillo, G. Induction of apoptosis in human cancer cells by candidaspongiolide, a novel sponge polyketide. J. Natl. Cancer Inst. 2008, 100, 1233–1246. [Google Scholar] [CrossRef] [PubMed]
- Philchenkov, A. Caspases: Potential targets for regulating cell death. J. Cell. Mol. Med. 2004, 8, 432–444. [Google Scholar] [CrossRef] [PubMed]
- Levine, B.; Kroemer, G. Autophagy in the pathogenesis of disease. Cell 2008, 132, 27–42. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Vicencio, J.M.; Kepp, O.; Tasdemir, E.; Maiuri, M.C.; Kroemer, G. To die or not to die: That is the autophagic qustion. Curr. Mol. Med. 2008, 8, 78–91. [Google Scholar] [PubMed]
- Jaeschke, H.; Lemasters, J.J. Apoptosis versus oncotic necrosis in hepatic ischemia/reperfusion injury. Gastroenterology 2003, 125, 1246–1257. [Google Scholar] [CrossRef]
- Stennicke, H.R.; Salvesen, G.S. Caspases-controlling intracellular signals byprotease zymogen activation. Biochim. Biophys. Acta 2000, 1477, 299–306. [Google Scholar] [CrossRef]
- Wang, J.Y.J. DNA damage and apoptosis. Cell Death Differ. 2001, 8, 1047–1048. [Google Scholar] [CrossRef] [PubMed]
- Lambert, L.A.; Qiao, N.; Hunt, K.K.; Lambert, D.H.; Mills, G.B.; Meijer, L.; Keyomarsi, K. Autophagy: A novel mechanism of synergistic cytotoxicity between doxorubicin and roscovitine in a sarcoma model. Cancer Res. 2008, 68, 7966–7974. [Google Scholar] [CrossRef] [PubMed]
- Rubinsztein, D.C.; Gestwicki, J.E.; Murphy, L.O.; Klionsky, D.D. Potential therapeutic applications of autophagy. Nat. Rev. Drug Discov. 2007, 6, 304–312. [Google Scholar] [CrossRef] [PubMed]
- Folmer, F.; Jaspars, M.; Dicato, M.; Diederich, M. Marine cytotoxins: Callers for the various dances of death. Gastroenterol. Hepatol. 2009, 2, 34–50. [Google Scholar]
- Degenhardt, K.; Mathew, R.; Beaudoin, B.; Bray, K.; Anderson, D.; Chen, G.; Mukherjee, C.; Shi, Y.; Gelinas, C.; Fan, Y.; et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell 2006, 10, 51–64. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.J.; Chee, C.E.; Huang, S.; Sinicrope, F.A. The role of autophagy in cancer: Therapeutic implications. Mol. Cancer Ther. 2011, 10, 1533–1541. [Google Scholar] [CrossRef] [PubMed]
- Yorimitsu, T.; Klionsky, D.J. Endoplasmic reticulum stress: A new pathway to induce autophagy. Autophagy 2007, 3, 160–662. [Google Scholar] [CrossRef] [PubMed]
- Thorburn, A. Apoptosis and autophagy: Regulatory connections between two supposedly different processes. Apoptosis 2008, 13, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Lum, J.J.; Bauer, D.E.; Kong, M.; Harris, M.H.; Li, C.; Lindsten, T.; Thompson, C.B. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell 2005, 120, 237–248. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N.; Levine, B. Autophagy in mammalian development and differentiation. Nat. Cell Biol. 2010, 12, 823–830. [Google Scholar] [CrossRef] [PubMed]
- Kroemer, G.; Marino, G.; Levine, B. Autophagy and the integrated stress response. Mol. Cell 2010, 40, 280–293. [Google Scholar] [CrossRef] [PubMed]
- Aravindan, S.; Ramraj, S.K.; Somasundaram, S.T.; Aravindan, N. Novel adjuvants from seaweed impede autophagy signaling in therapy-resistant residual pancreatic cancer. J. Biomed. Sci. 2015, 22, 28. [Google Scholar] [CrossRef] [PubMed]
- Shintani, T.; Klionsky, D.J. Autophagy in health and disease: A double-edged sword. Science 2004, 306, 990–995. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N.; Levine, B.; Cuervo, A.M.; Klionsky, D.J. Autophagy fights disease through cellular self-digestion. Nature 2008, 451, 1069–1075. [Google Scholar] [CrossRef] [PubMed]
- Virgin, H.W.; Levine, B. Autophagy genes in immunity. Nat. Immunol. 2009, 10, 461–470. [Google Scholar] [CrossRef] [PubMed]
- Levine, B.; Mizushima, N.; Virgin, H.W. Autophagy in immunity and inflammation. Nature 2011, 469, 323–335. [Google Scholar] [CrossRef] [PubMed]
- Rabinowitz, J.D.; White, E. Autophagy and metabolism. Science 2010, 330, 1344–1348. [Google Scholar] [CrossRef] [PubMed]
- Pavlides, S.; Tsirigos, A.; Vera, I.; Flomenberg, N.; Frank, P.G.; Casimiro, M.C. Loss of stromal caveolin-1 leads to oxidative stress, mimics hypoxia and drives inflammation in the tumor microenvironment, conferring the “Reverse Warburg Effect”: A transcriptional informatics analysis with validation. Cell Cycle 2010, 9, 2201–2219. [Google Scholar] [CrossRef] [PubMed]
- Hippert, M.M.; O’Toole, P.S.; Thorburn, A. Autophagy in cancer: Good, bad, or both? Cancer Res. 2006, 66, 9349–9351. [Google Scholar] [CrossRef] [PubMed]
- Kimmelman, A.C. The dynamic nature of autophagy in cancer. Genes Dev. 2011, 25, 1999–2010. [Google Scholar] [CrossRef] [PubMed]
- Kondo, Y.; Kanzawa, T.; Sawaya, R.; Kondo, S. The role of autophagy in cancer development and response to therapy. Nat. Rev. Cancer 2005, 5, 726–734. [Google Scholar] [CrossRef] [PubMed]
- Mathew, R.; Karantza-Wadsworth, V.; White, E. Role of autophagy in cancer. Nat. Rev. Cancer 2007, 7, 961–967. [Google Scholar] [CrossRef] [PubMed]
- Altman, B.J.; Rathmell, J.C. Metabolic stress in autophagy and cell death pathways. Cold Spring Harb. Perspect. Biol. 2012, 4, a008763. [Google Scholar] [CrossRef] [PubMed]
- Qu, X.; Yu, J.; Bhagat, G.; Furuya, N.; Hibshoosh, H.; Troxel, A.; Rosen, J.; Eskelinen, E.L.; Mizushima, N.; Ohsumi, Y.; et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J. Clin. Investig. 2003, 112, 1809–1820. [Google Scholar] [CrossRef] [PubMed]
- Pattingre, S.; Espert, L.; Biard-Piechaczyk, M.; Codogno, P. Regulation of macroautophagy by mTOR and Beclin 1 complexes. Biochimie 2008, 90, 313–323. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, H.; Kondo, Y.; Fujiwara, K.; Kanzawa, T.; Aoki, H.; Mills, G.B.; Kondo, S. Synergistic augmentation of rapamycin-induced autophagy in malignant glioma cells by phosphatidylinositol 3-kinase/protein kinase B inhibitors. Cancer Res. 2005, 65, 3336–3346. [Google Scholar] [PubMed]
- Semenza, G.L. Defining the role of hypoxia-inducible factor 1 in cancer biology and therapeutics. Oncogene 2010, 29, 625–634. [Google Scholar] [CrossRef] [PubMed]
- Jung, C.H.; Jun, C.B.; Ro, S.H.; Kim, Y.M.; Otto, N.M.; Cao, J.; Kundu, M.; Kim, D.H. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol. Biol. Cell 2009, 20, 1992–2003. [Google Scholar] [CrossRef] [PubMed]
- Maiuri, M.C.; Malik, S.A.; Morselli, E.; Kepp, O.; Criollo, A.; Mouchel, P.L.; Carnuccio, R.; Kroemer, G. Stimulation of autophagy by the p53 target gene Sestrin2. Cell Cycle 2009, 8, 1571–1576. [Google Scholar] [CrossRef] [PubMed]
- White, E.; Di Paola, R.S. The double-edged sword of autophagy modulation in cancer. Clin. Cancer Res. 2009, 15, 5308–5316. [Google Scholar] [CrossRef] [PubMed]
- Mathew, R.; Karp, C.M.; Beaudoin, B.; Vuong, N.; Chen, G.; Chen, H.Y.; Bray, K.; Reddy, A.; Bhanot, G.; Gelinas, C.; et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell 2009, 137, 1062–1075. [Google Scholar] [CrossRef] [PubMed]
- Semenza, G.L. Regulation of cancer cell metabolism by hypoxia-inducible factor 1. Semin. Cancer Biol. 2009, 19, 12–16. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.H.; Jackson, S.; Seaman, M.; Brown, K.; Kempkes, B.; Hibshoosh, H.; Levine, B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 1999, 402, 672–676. [Google Scholar] [PubMed]
- Tang, D.; Kang, R.; Livesey, K.M.; Cheh, C.W.; Farkas, A.; Loughran, P. Endogenous HMGB1 regulates autophagy. J. Cell Biol. 2010, 190, 881–892. [Google Scholar] [CrossRef] [PubMed]
- Kanzawa, T.; Zhang, L.; Xiao, L.; Germano, I.M.; Kondo, Y.; Kondo, S. Arsenic trioxide induces autophagic cell death in malignant glioma cells by upregulation of mitochondrial cell death protein BNIP3. Oncogene 2005, 24, 980–991. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Gao, Z.; Marks, P.A.; Jiang, X. Apoptotic and autophagic cell death induced by histone deacetylase inhibitors. Proc. Natl. Acad. Sci. USA 2004, 101, 18030–18035. [Google Scholar] [CrossRef] [PubMed]
- Turcotte, S.; Chan, D.A.; Sutphin, P.D.; Hay, M.P.; Denny, W.A.; Giaccia, A.J. A molecule targeting vhl-deficient renal cell carcinoma that induces autophagy. Cancer Cell 2008, 14, 90–102. [Google Scholar] [CrossRef] [PubMed]
- Gewirtz, D.A. The four faces of autophagy: Implications for cancer therapy. Cancer Res. 2014, 74, 647–651. [Google Scholar] [CrossRef] [PubMed]
- Chung, C.; Mader, C.C.; Schmitz, J.C.; Atladottir, J.; Fitchev, P.; Cornwell, M.L.; Koleske, A.J.; Crawford, S.E.; Gorelick, F. The vacuolar-ATPase modulates matrix metalloproteinase isoforms in human pancreatic cancer. Lab. Investig. 2011, 91, 732–743. [Google Scholar] [CrossRef] [PubMed]
- Fais, S.; De Milito, A.; You, H.; Qin, W. Targeting vacuolar H+-ATPases as a new strategy against cancer. Cancer Res. 2007, 67, 10627–10630. [Google Scholar] [CrossRef] [PubMed]
- Sennoune, S.R.; Luo, D.; Martinez-Zaguilan, R. Plasmalemmal vacuolar-type H+-ATPase in cancer biology. Cell Biochem. Biophys. 2004, 40, 185–206. [Google Scholar] [CrossRef]
- Spugnini, E.P.; Citro, G.; Fais, S. Proton pump inhibitors as anti vacuolar-ATPases drugs: A novel anticancer strategy. J. Exp. Clin. Cancer Res. 2010, 29, 44. [Google Scholar] [CrossRef] [PubMed]
- Kallifatidis, G.; Hoepfner, D.; Jaeg, T.; Guzmán, E.A.; Wright, A.E. The Marine Natural Product Manzamine A Targets Vacuolar ATPases and Inhibits Autophagy in Pancreatic Cancer Cells. Mar. Drugs 2013, 11, 3500–3516. [Google Scholar] [CrossRef] [PubMed]
- Aghajan, M.; Li, N.; Karin, M. Obesity, autophagy and the pathogenesis of liver and pancreatic cancers. J. Gastroenterol. Hepatol. 2012, 27 (Suppl. S2), 10–14. [Google Scholar] [CrossRef] [PubMed]
- Maiuri, M.C.; Zalckvar, E.; Kimchi, A.; Kroemer, G. Self-eating and self-killing: Crosstalk between autophagy and apoptosis. Nat. Rev. Mol. Cell Biol. 2007, 8, 741–752. [Google Scholar] [CrossRef] [PubMed]
- Bursch, W.; Ellinger, A.; Kienzl, H.; Torok, L.; Pandey, S.; Hermann, R. Active cell death induced by the anti-estrogens tamoxifen and ICI 164 384 in human mammary carcinoma cells (MCF-7) in culture: The role of autophagy. Carcinogen 1996, 17, 1595–1607. [Google Scholar] [CrossRef]
- Jin, Z.; Li, Y.; Pitti, R.; Lawrence, D.; Pham, V.C.; Lill, J.R. Cullin3-based polyubiquitination and p62-dependent aggregation of caspase-8 mediate extrinsic apoptosis signaling. Cell 2009, 137, 721–735. [Google Scholar] [CrossRef] [PubMed]
- Levine, B.; Sinha, S.; Kroemer, G. Bcl-2 family members: Dual regulators of apoptosis and autophagy. Autophagy 2008, 4, 600–606. [Google Scholar] [CrossRef] [PubMed]
- Cragg, G.M.; Grothaus, P.G.; Newman, D.J. Impact of natural products on developing new anti-cancer agents. Chem. Rev. 2009, 109, 3012–3043. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Z.Q.; Zhang, Z.P.; Li, J.H.; Wei, S.J.; Tu, G.Q. Characterization of Streptomyces padanus JAU4234, a producer of actinomycin X2, fungichromin, and a new polyene macrolide antibiotic. Appl. Environ. Microbiol. 2012, 78, 589–592. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Z.Q.; Wang, J.F.; Hao, Y.Y.; Wang, Y. Recent advances in the discovery and development of marine microbial natural products. Mar. Drugs 2013, 11, 700–717. [Google Scholar] [CrossRef] [PubMed]
- Blunt, J.W.; Copp, B.R.; Keyzers, R.A.; Munroa, M.H.G.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2015, 32, 116–211. [Google Scholar] [CrossRef] [PubMed]
- Proksch, P.; Edrada, R.A.; Ebel, R. Drugs from the seas-current status and microbiological implications. Appl. Microbiol. Biotechnol. 2002, 59, 125–134. [Google Scholar] [PubMed]
- Dyshlovoy, S.A.; Hauschild, J.; Amann, K.; Tabakmakher, K.M.; Venz, S.; Walther, R.; Guzii, A.G.; Makarieva, T.N.; Shubina, L.K.; Fedorov, S.N.; et al. Marine alkaloid Monanchocidin a overcomes drug resistance by induction of autophagy and lysosomal membrane permeabilization. Oncotarget 2015, 6, 17328–17341. [Google Scholar] [CrossRef] [PubMed]
- Molinski, T.F.; Dalisay, D.S.; Lievens, S.L.; Saludes, J.P. Drug development from marine natural products. Nat. Rev. Drug Discov. 2009, 8, 69–85. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.J.; Cragg, G.M. Marine natural products and related compounds in clinical and advanced preclinical trials. J. Nat. Prod. 2004, 67, 1216–1238. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.J.; Cragg, G.M. Marine-sourced anti-cancer and cancer pain control agents in clinical and late preclinical development. Mar. Drugs 2014, 12, 255–278. [Google Scholar] [CrossRef] [PubMed]
- Stonik, V. Marine natural products: A way to new drugs. Acta Naturae 2009, 2, 15–25. [Google Scholar]
- Galmarini, C.M.; D’Incalci, M.; Allavena, P. Trabectedin and plitidepsin: Drugs from the sea that strike the tumor microenvironment. Mar. Drugs 2014, 12, 719–733. [Google Scholar] [CrossRef] [PubMed]
- Mehbub, M.F.; Lei, J.; Franco, C.; Zhang, W. Marine sponge derived natural products between 2001 and 2010: Trends and opportunities for discovery of bioactives. Mar. Drugs 2014, 12, 4539–4577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fusetani, N. Marine toxins: An overview. Prog. Mol. Subcell. Biol. 2009, 46, 1–44. [Google Scholar] [PubMed]
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs over the 30 Years from 1981 to 2010. J. Nat. Prod. 2012, 75, 311–335. [Google Scholar] [CrossRef] [PubMed]
- Mayer, A.M.; Gustafson, K.R. Marine pharmacology in 2000: Antitumor and cytotoxic compounds. Int. J. Cancer 2003, 105, 291–299. [Google Scholar] [CrossRef] [PubMed]
- Mayer, A.M.; Gustafson, K.R. Marine pharmacology in 2001–2012: Antitumour and cytotoxic compounds. Eur. J. Cancer 2004, 40, 2676–2704. [Google Scholar] [CrossRef] [PubMed]
- Mayer, A.M.; Gustafson, K.R. Marine pharmacology in 2003–2004: Anti-tumour and cytotoxic compounds. Eur. J. Cancer 2006, 42, 2241–2270. [Google Scholar] [CrossRef] [PubMed]
- Vega Thurber, R.; Epel, D. Apoptosis in early development of the sea urchin, Strongylocentrotus purpuratus. Dev. Biol. 2007, 303, 336–346. [Google Scholar] [CrossRef] [PubMed]
- Shimada, M.; Satoh, N.; Yokosawa, H. Involvement of Rel/NF-kappaB in regulation of ascidian notochord formation. Dev. Growth Diff. 2001, 43, 145–154. [Google Scholar] [CrossRef]
- James, E.R.; Green, D.R. Manipulation of apoptosis in the host-parasite interaction. Trends Parasitol. 2004, 20, 280–287. [Google Scholar] [CrossRef] [PubMed]
- Moore, M.N.; Viarengo, A.; Donkin, P.; Hawkins, A.J.S. Autophagic and lysosomal reactions to stress in the hepatopancreas of blue mussels. Aquat. Toxicol. 2007, 84, 80–91. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Zheng, J.; Zhang, Z.; Dong, X.; Zhao, L.; Tada, M. Autophagy plays a potential role in the process of sea cucumber body wall “melting” induced by UV irradiation. Wuhan Univ. J. Nat. Sci. 2008, 13, 232–238. [Google Scholar] [CrossRef]
- Dunn, S.R.; Schnitzler, C.E.; Weis, V.M. Apoptosis and autophagy as mechanisms of dinoflagellate symbiont release during cnidarian bleaching: Every which way you lose. Proc. R. Soc. B Biol. Sci. 2007, 274, 3079–3085. [Google Scholar] [CrossRef] [PubMed]
- Farooqi, A.A.; Yaylim, I.; Ozkan, N.E.; Zaman, F.; Halim, T.A.; Chang, H.W. Restoring trail mediated signaling in ovarian cancer cells. Arch. Immunol. Ther. Exp. (Warsz.) 2014, 62, 459–474. [Google Scholar] [CrossRef] [PubMed]
- Talero, E.; García-Mauriño, S.; Ávila-Román, J.; Rodríguez-Luna, A.; Alcaide, A.; Motilva, V. Bioactive compounds isolated from microalgae in chronic inflammation and cancer. Mar. Drugs 2015, 13, 6152–6209. [Google Scholar] [CrossRef] [PubMed]
- Martins, A.; Vieira, H.; Gaspar, H.; Santos, S. Marketed marine natural products in the pharmaceutical and cosmeceutical industries: Tip of success. Mar. Drugs 2015, 12, 1066–1101. [Google Scholar] [CrossRef] [PubMed]
- Datta, D.; Talapatra, S.N.; Swarnakar, S. Bioactive compounds from marine invertebrates for potential medicines—An overview. Int. Lett. Nat. Sci. 2015, 34, 42–61. [Google Scholar] [CrossRef]
- Forestieri, R.; Donohue, E.; Balgi, A.; Roberge, M.; Andersen, R.J. Synthesis of clionamine B, an autophagy stimulating aminosteroid isolated from the sponge Cliona celata. Org. Lett. 2013, 15, 3918–3921. [Google Scholar] [CrossRef] [PubMed]
- Lam, K.K.Y.; Zheng, X.; Forestieri, R.; Balgi, A.D.; Nodwell, M.; Vollett, S.; Anderson, H.J.; Andersen, R.J.; Av-Gay, Y.; Roberge, M. Nitazoxanide stimulates autophagy and inhibits mTORC1 signaling and intracellular proliferation of Mycobacterium tuberculosis. PLoS Pathog. 2012, 8, e10022691. [Google Scholar] [CrossRef] [PubMed]
- Keyzers, R.A.; Daoust, J.; Davies-Coleman, M.T.; Van Soest, R.; Balgi, A.; Donohue, E.; Roberge, M.; Andersen, R.J. Autophagy-modulating aminosteroids isolated from the sponge Cliona celata. Org. Lett. 2008, 10, 2959–2962. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, M.; Endo, M.; Tanaka, N.; Gen-Pei, L. Structures of xestospongin A,B,C and D, novel vasodilative compounds from marine sponge, Xestospongia exigua. Tetrahedron Lett. 1984, 25, 3227–3230. [Google Scholar] [CrossRef]
- Jaimovich, E.; Mattei, C.; Liberona, J.L.; Cardenas, C.; Estrada, M.; Barbier, J.; Debitus, C.; Laurent, D.; Molgó, J. Xestospongin B, a competitive inhibitor of IP3-mediated Ca2+ signalling in cultured rat myotubes, isolated myonuclei, and neuroblastoma (NG108–15) cells. FEBS Lett. 2005, 579, 2051–2057. [Google Scholar] [CrossRef] [PubMed]
- Akl, M.R.; Ayoub, N.M.; Ebrahim, H.Y.; Mohyeldin, M.M.; Orabi, K.Y.; Foudah, A.I.; El Sayed, K. Araguspongine C induces autophagic death in breast cnacer cells through suppression of c-Met and HER2 receptor tyrosine in kinase signaling. Mar. Drugs 2015, 13, 288–311. [Google Scholar] [CrossRef] [PubMed]
- Guzii, A.G.; Makarieva, T.N.; Denisenko, V.A.; Dmitrenok, P.S.; Kuzmich, A.S.; Dyshlovoy, S.A.; Krasokhin, V.B.; Stonik, V.A. Monanchocidin: A new apoptosis-inducing polycyclic guanidine alkaloid from the marine sponge Monanchora pulchra. Org. Lett. 2010, 12, 4292–4295. [Google Scholar] [CrossRef] [PubMed]
- Seglen, P.O.; Gordon, P.B. 3-Methyladenine: Specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc. Natl. Acad. Sci. USA 1982, 79, 1889–1892. [Google Scholar] [CrossRef] [PubMed]
- Kanno, S.; Yomogida, S.; Tomizawa, A.; Yamazaki, H.; Ukai, K.; Mangindaan, R.E.; Namikoshi, M.; Ishikawa, M. Papuamine causes autophagy following the reduction of cell survival through mitochondrial damage and JNK activation in MCF-7 human breast cancer cells. Int. J. Oncol. 2013, 43, 1413–1419. [Google Scholar] [CrossRef] [PubMed]
- Su, J.Y.; Meng, Y.H.; Zeng, L.M.; Fu, X.; Schmitz, F.J. Stellettin A, a new triterpenoid pigment from the marine sponge Stelletta tenuis. J. Nat. Prod. 1994, 57, 1450–1451. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.K.; Ling, Y.H.; Cheung, F.W.; Che, C.T. Stellettin A induces endoplasmic reticulum stress in murine B16 melanoma cells. J. Nat. Prod. 2012, 75, 586–590. [Google Scholar] [CrossRef] [PubMed]
- Li, D.D.; Guo, J.F.; Huang, J.J.; Wang, L.L.; Deng, R.; Liu, J.N.; Feng, G.K.; Xiao, D.J.; Deng, S.Z.; Zhang, X.S.; et al. Rhabdastrellic acid-A induced autophagy-associated cell death through blocking Akt pathway in human cancer cells. PLoS ONE 2010, 5, e12176. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-Y.; Chung, K.J.; Hwang, I.H.; Gwak, J.; Park, S.; Ju, B.G.; Yun, E.; Kim, D.-E.; Chung, Y.-H.; Na, M.; et al. Activation of p53 with Ilimaquinone and ethylsmenoquinine, marine sponge metabolites, indices apoptosis and autophagy in colon cancer cells. Mar. Drugs 2015, 13, 543–557. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.; Kundu, R. Antiproliferative activity of methanolic extracts from two green algae, Enteromorpha intestinalis and Rhizoclonium riparium on HeLa cells. DARU 2013, 21. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, A.O.; Veiga-Santos, P.; Ueda-Nakamura, T.; Filho, B.P.; Sudatti, D.B.; Bianco, E.M.; Pereira, R.C.; Nakamura, C.V. Effect of elatol, isolated from red seaweed Laurencia dendroidea, on Leishmania amazonensis. Mar. Drugs 2010, 8, 2733–2743. [Google Scholar] [CrossRef] [PubMed]
- Hou, L.L.; Gao, C.; Chen, L.; Hu, G.Q.; Xie, S.Q. Essential role of autophagy in fucoxanthin-induced cytotoxicity to human epithelial cervical cancer HeLa cells. Acta Pharmacol. Sin. 2013, 34, 1403–1410. [Google Scholar] [CrossRef] [PubMed]
- Medina, R.A.; Goeger, D.E.; Hills, P.; Mooberry, S.L.; Huang, N.; Romero, L.I.; Ortega-Barría, E.; Gerwick, W.H.; McPhail, K.L. Coibamide A, a potent antiproliferative cyclic depsipeptide from the Panamanian marine cyanobacterium Leptolyngbya sp. J. Am. Chem. Soc. 2008, 130, 6324–6325. [Google Scholar] [CrossRef] [PubMed]
- Hau, A.M.; Greenwood, J.A.; Lohr, C.V.; Serrill, J.D.; Proteau, P.J.; Ganley, I.G.; McPhail, K.L.; Ishmael, J.E. Coibamide A induces mTOR-independent autophagy and cell death in human glioblastoma cells. PLoS ONE 2013, 8, e65250. [Google Scholar] [CrossRef] [PubMed]
- Russo, G.L.; Russo, M.; Castellano, I.; Napolitano, A.; Palumbo, A. Ovothiol isolated from sea urchin oocytes induces autophagy in the Hep-G2 cell line. Mar. Drugs 2014, 12, 4069–4085. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.Y.S.; Abrell, L.M.; Avelar, A.; Borgeson, B.M.; Crews, P. New hirsutane based sesquiquiterpenes from salt water cultures of a marine sponge-derived fungus and the terrestrial fungus Coriolus consors. Tetrahedron 1998, 54, 7335–7342. [Google Scholar] [CrossRef]
- Yang, F.; Chen, W.D.; Deng, R.; Zhang, H.; Tang, J.; Wu, K.W.; Li, D.D.; Feng, G.K.; Lan, W.J.; Li, H.J.; et al. Hirsutanol A, a novel sesquiterpene compound from fungus Chondrostereum sp., induces apoptosis and inhibits tumor growth through mitochondrial-independent ROS production: Hirsutanol A inhibits tumor growth through ROS production. J. Transl. Med. 2013, 11, 32. [Google Scholar] [CrossRef] [PubMed]
- Zhu, K.; Dunner, K., Jr.; McConkey, D.J. Proteasome inhibitors activate autophagy as a cytoprotective response in human prostate cancer cells. Oncogene 2010, 29, 451–462. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Chen, H.; Shang, Z.; Jiao, B.; Yuan, B.; Sun, W.; Wang, B.; Miao, M.; Huang, C. SD118-xanthocillin X (1), a novel marine agent extracted from Penicillium commune, induces autophagy through the inhibition of the MEK/ERK pathway. Mar. Drugs 2012, 10, 1345–1359. [Google Scholar] [CrossRef] [PubMed]
- Guimaraes, L.A.; Jimenez, P.C.; da Silva Sousa, T.; Freitas, H.P.S.; Rocha, D.D.; Wilke, D.V.; Martin, J.; Reves, F.; Pessoa, O.D.L.; Costa-Lotufo, L.V. Chromomycin A2 induces autophagy in melanoma cells. Mar. Drugs 2014, 12, 5839–5855. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, Q.H.; Zhang, X.C.; Fu, T.; Gu, J.Z.; Wang, L.; Wang, Y.; Lai, Y.B.; Wang, Y.Q.; Guo, Y. Omega-3 polyunsaturated fatty acids inhibit the proliferation of the lung adenocarcinoma cell line A549 in vitro. Mol. Med. Rep. 2014, 9, 401–406. [Google Scholar] [PubMed]
- Ivanchina, N.V.; Kicha, A.A.; Stonik, V.A. Steroid glycosides from marine organisms. Steroids 2011, 76, 425–454. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhan, Y.; Xu, R.; Shao, R.; Jiang, J.; Wang, Z. Src mediates extracellular signal-regulated kinase 1/2 activation and autophagic cell death induced by cardiac glycosides in human non-small cell lung cancer cell lines. Mol. Carcinog. 2015, 54, E26–E34. [Google Scholar] [CrossRef] [PubMed]
- Edrada, R.A.; Proksch, P.; Wray, V.; Witte, L.; Muller, W.E.; Van Soest, R.W. Four new bioactive manzamine-type alkaloids from the Philippine marine sponge Xestospongia ashmorica. J. Nat. Prod. 1996, 59, 1056–1060. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, D.; Tsuda, M.; Kobayashi, J. Three new manzamine congeners from amphimedon sponge. J. Nat. Prod. 1998, 61, 689–692. [Google Scholar] [CrossRef] [PubMed]
- Ichiba, T.; Corgiat, J.M.; Scheuer, P.J.; Kelly-Borges, M. 8-Hydroxymanzamine A, a beta-carboline alkaloid from a sponge, Pachypellina sp. J. Nat. Prod. 1994, 57, 168–170. [Google Scholar] [CrossRef] [PubMed]
- Ang, K.K.; Holmes, M.J.; Kara, U.A. Immune-mediated parasite clearance in mice infected with Plasmodium berghei following treatment with manzamine A. Parasitol. Res. 2001, 87, 715–721. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Hu, J.F.; Kazi, A.B.; Li, Z.; Avery, M.; Peraud, O.; Hill, R.T.; Franzblau, S.G.; Zhang, F.; Schinazi, R.F.; et al. Manadomanzamines A and B: A novel alkaloid ring system with potent activity against mycobacteria and HIV-1. J. Am. Chem. Soc. 2003, 125, 13382–13386. [Google Scholar] [CrossRef] [PubMed]
- Rao, K.V.; Santarsiero, B.D.; Mesecar, A.D.; Schinazi, R.F.; Tekwani, B.L.; Hamann, M.T. New manzamine alkaloids with activity against infectious and tropical parasitic diseases from an Indonesian sponge. J. Nat. Prod. 2003, 66, 823–828. [Google Scholar] [CrossRef] [PubMed]
- Rao, K.V.; Kasanah, N.; Wahyuono, S.; Tekwani, B.L.; Schinazi, R.F.; Hamann, M.T. Three new manzamine alkaloids from a common Indonesian sponge and their activity against infectious and tropical parasitic diseases. J. Nat. Prod. 2004, 67, 1314–1318. [Google Scholar] [CrossRef] [PubMed]
- Yousaf, M.; Hammond, N.L.; Peng, J.; Wahyuono, S.; McIntosh, K.A.; Charman, W.N.; Mayer, A.M.; Hamann, M.T. New manzamine alkaloids from an Indo-Pacific sponge. Pharmacokinetics, oral availability, and the significant activity of several manzamines against HIV-I, AIDS opportunistic infections, and inflammatory diseases. J. Med. Chem. 2004, 47, 3512–3517. [Google Scholar] [CrossRef] [PubMed]
- Guzman, E.A.; Johnson, J.D.; Linley, P.A.; Gunasekera, S.E.; Wright, A.E. A novel activity from an old compound: Manzamine A reduces the metastatic potential of AsPC-1 pancreatic cancer cells and sensitizes them to TRAIL-induced apoptosis. Investig. New Drugs 2011, 29, 777–785. [Google Scholar] [CrossRef] [PubMed]
- Randazzo, A.; Debitus, C.; Minale, L.; Garcia Pastor, P.; Alcaraz, M.J.; Paya, M.; Gomez-Paloma, L. Petrosaspongiolides M–R: New potent and selective phospholipase A2 inhibitors from the New Caledonian marine sponge Petrosaspongia nigra. J. Nat. Prod. 1998, 61, 571–575. [Google Scholar] [CrossRef] [PubMed]
- Monti, M.C.; Margarucci, L.; Riccio, R.; Bonfili, L.; Mozzicafreddo, M.; Eleuteri, A.M.; Casapullo, A. Mechanistic insights on petrosaspongiolide M inhibitory effects on immunoproteasome and autophagy. Biochim. Biophys. Acta 2014, 1844, 713–721. [Google Scholar] [CrossRef] [PubMed]
- Carr, G.; Williams, D.E.; Diaz-Marrero, A.R.; Patrick, B.O.; Bottriell, H.; Balgi, A.D.; Donohue, E.; Roberge, M.; Andersen, R.J. Bafilomycins produced in culture by Streptomyces spp. isolated from marine habitats are potent inhibitors of autophagy. J. Nat. Prod. 2010, 73, 422–427. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.; Cai, J.; Sun, L.; Li, Y.; Qu, J.; Snider, B.J.; Wu, S. Proteasome inhibitors activate autophagy involving inhibition of PI3K-Akt-mTOR pathway as an anti-oxidation defense in human RPE cells. PLoS ONE 2014, 9, e103364. [Google Scholar] [CrossRef] [PubMed]
- Yoshie, Y.; Wei, W.; Ya-Pei, H.; Takeshi, S. Compositional difference of phenolic compounds between two seaweeds, Halimeda spp. J. Tokyo Univ. Fish. 2002, 88, 21–24. [Google Scholar]
- Furusawa, E.; Furusawa, S. Anticancer activity of a natural product, viva-natural, extracted from Undaria pinnantifida on intraperitoneally implanted Lewis lung carcinoma. Oncology 1985, 42, 364–369. [Google Scholar] [CrossRef] [PubMed]
- Aravindan, S.; Delma, C.R.; Thirugnanasambandan, S.S.; Herman, T.S.; Aravindan, N. Anti-pancreatic cancer deliverables from sea: First-hand evidence on the efficacy, molecular targets and mode of action for multifarious polyphenols from five different brown-algae. PLoS ONE 2013, 8, e61977. [Google Scholar] [CrossRef] [PubMed]





| Compound | Source | Structure | Autophagy Print | Disease |
|---|---|---|---|---|
| Monanchocidin A | sponge | alkaloid | Inducer | Leukemia, Prostate Cancer |
| Clionamines | sponge | aminosteroid | Inducer | Breast cancer |
| Papuamine | sponge | alkaloid | Inducer | Breast cancer |
| Rhabdastrellic acid A | sponge | triterpenoid | Inducer | Lung cancer |
| Stellettin A | sponge | triterpene | Inducer | Melanoma |
| Xestospongin B | sponge | alkaloid | Inducer | Neuroblastoma |
| Araguspongine C | sponge | alkaloid | Inducer | Breast cancer |
| Ilamaquinone | sponge | quinone | Inducer | Colon cancer |
| Ovothiol A | sea urchin | thiol | Inducer | Liver cancer |
| Hirsutanol | fungus | sesquiterpene | Inducer | Breast cancer |
| Xanthocillin X | fungus | diphenol | Inducer | Liver cancer |
| Salinosporamide A | bacteria | lactone | Inducer | Prostate Cancer |
| Chromomycin A2 | bacteria | polyketide | Inducer | Melanoma |
| Coibamide A | cyanobacterium | cyclopeptide | Inducer | Glioblastoma |
| EPA and DHA | algae | fatty acids | Inducer | Lung cancer |
| Fucoxanthin | algae | carotenoid | Inducer | Uterine Cancer |
| Methanolic extract | algae | phenol | Inducer | Uterine Cancer |
| Manzamine A | sponge | alkaloid | Inhibitor | Pancreatic cancer |
| Petrosaspongiolide | sponge | terpenoid | Inhibitor | Chronic inflammation |
| Bafilomycin | algae | macrolide antibiotic | Inhibitor | Retinal disease |
| Polyphenols | algae | phenol | Inhibitor | Pancreatic cancer |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruocco, N.; Costantini, S.; Costantini, M. Blue-Print Autophagy: Potential for Cancer Treatment. Mar. Drugs 2016, 14, 138. https://doi.org/10.3390/md14070138
Ruocco N, Costantini S, Costantini M. Blue-Print Autophagy: Potential for Cancer Treatment. Marine Drugs. 2016; 14(7):138. https://doi.org/10.3390/md14070138
Chicago/Turabian StyleRuocco, Nadia, Susan Costantini, and Maria Costantini. 2016. "Blue-Print Autophagy: Potential for Cancer Treatment" Marine Drugs 14, no. 7: 138. https://doi.org/10.3390/md14070138
APA StyleRuocco, N., Costantini, S., & Costantini, M. (2016). Blue-Print Autophagy: Potential for Cancer Treatment. Marine Drugs, 14(7), 138. https://doi.org/10.3390/md14070138