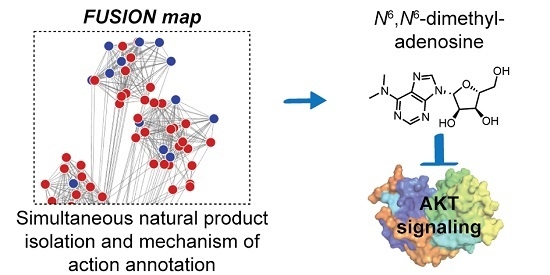
FUSION-Guided Hypothesis Development Leads to the Identification of N6,N6-Dimethyladenosine, a Marine-Derived AKT Pathway Inhibitor
Abstract
:1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. Collection and Phylogenetic Analysis of Strain SNA-024
4.2. Cultivation and Extraction
4.3. Purification of a Fraction Enriched in N6,N6-Dimethyladenosine
4.4. NMR Characterization
4.5. Reagents and Antibodies
4.6. In-Cell Western
4.7. Reverse Phase Protein Array
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Wolfender, J.-L.; Marti, G.; Thomas, A.; Bertrand, S. Current approaches and challenges for the metabolite profiling of complex natural extracts. J. Chromatogr. A 2015, 1382, 136–164. [Google Scholar] [CrossRef] [PubMed]
- Potts, M.B.; Kim, H.S.; Fisher, K.W.; Hu, Y.; Carrasco, Y.P.; Bulut, G.B.; Ou, Y.-H.; Herrera-Herrera, M.L.; Cubillos, F.; Mendiratta, S.; et al. Using functional signature ontology (FUSION) to identify mechanisms of action for natural products. Sci. Signal. 2013, 6, ra90. [Google Scholar] [CrossRef] [PubMed]
- Capodanno, A.; Camerini, A.; Orlandini, C.; Baldini, E.; Resta, M.L.; Bevilacqua, G.; Collecchi, P. Dysregulated PI3K/Akt/PTEN pathway is a marker of a short disease-free survival in node-negative breast carcinoma. Hum. Pathol. 2009, 40, 1408–1417. [Google Scholar] [CrossRef] [PubMed]
- Vivanco, I.; Chen, Z.C.; Tanos, B.; Oldrini, B.; Hsieh, W.-Y.; Yannuzzi, N.; Campos, C.; Mellinghoff, I.K. A kinase-independent function of AKT promotes cancer cell survival. eLife 2014, 3, e03751. [Google Scholar] [CrossRef] [PubMed]
- Sanidas, I.; Polytarchou, C.; Hatziapostolou, M.; Ezell, S.A.; Kottakis, F.; Hu, L.; Guo, A.; Xie, J.; Comb, M.J.; Iliopoulos, D.; et al. Phosphoproteomics screen reveals Akt isoform-specific signals linking RNA processing to lung cancer. Mol. Cell 2014, 53, 577–590. [Google Scholar] [CrossRef] [PubMed]
- Ye, Q.; Cai, W.; Zheng, Y.; Evers, B.M.; She, Q.B. ERK and AKT signaling cooperate to translationally regulate survivin expression for metastatic progression of colorectal cancer. Oncogene 2014, 33, 1828–1839. [Google Scholar] [CrossRef] [PubMed]
- Yue, S.; Li, J.; Lee, S.-Y.; Lee, H.J.; Shao, T.; Song, B.; Cheng, L.; Masterson, T.A.; Liu, X.; Ratliff, T.L.; et al. Cholesteryl ester accumulation induced by PTEN loss and PI3K/AKT activation underlies human prostate cancer aggressiveness. Cell Metab. 2014, 19, 393–406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ocana, A.; Vera-Badillo, F.; Al-Mubarak, M.; Templeton, A.J.; Corrales-Sanchez, V.; Diez-Gonzalez, L.; Cuenca-Lopez, M.D.; Seruga, B.; Pandiella, A.; Amir, E. Activation of the PI3K/mTOR/AKT pathway and survival in solid tumors: Systematic review and meta-analysis. PLoS ONE 2014, 9, e95219. [Google Scholar] [CrossRef] [PubMed]
- Ou, Y.-H.; Torres, M.; Ram, R.; Formstecher, E.; Roland, C.; Cheng, T.; Brekken, R.; Wurz, R.; Tasker, A.; Polverino, T.; et al. TBK1 directly engages Akt/PKB survival signaling to support oncogenic transformation. Mol. Cell 2011, 41, 458–470. [Google Scholar] [CrossRef] [PubMed]
- Alessi, D.R.; James, S.R.; Downes, C.P.; Holmes, A.B.; Gaffney, P.R.J.; Reese, C.B.; Cohen, P. Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Bα. Curr. Biol. 1997, 7, 261–269. [Google Scholar] [CrossRef]
- Yu, J.; Zhou, X.; Chang, M.; Nakaya, M.; Chang, J.-H.; Xiao, Y.; William Lindsey, J.; Dorta-Estremera, S.; Cao, W.; Zal, A.; et al. Regulation of T-cell activation and migration by the kinase TBK1 during neuroinflammation. Nat. Commun. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Limbach, P.A.; Crain, P.F.; McCloskey, J.A. Summary: The modified nucleosides of RNA. Nucleic Acids Res. 1994, 22, 2183–2196. [Google Scholar] [CrossRef] [PubMed]
- Marbaniang, C.N.; Vogel, J. Emerging roles of RNA modifications in bacteria. Curr. Opin. Microbiol. 2016, 30, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Machnicka, M.A.; Milanowska, K.; Osman Oglou, O.; Purta, E.; Kurkowska, M.; Olchowik, A.; Januszewski, W.; Kalinowski, S.; Dunin-Horkawicz, S.; Rother, K.M.; et al. Modomics: A database of RNA modification pathways—2013 update. Nucleic Acids Res. 2013, 41, D262–D267. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.T.; Chionh, Y.H.; Ho, C.H.; Lim, K.S.; Babu, I.R.; Ang, E.; Wenwei, L.; Alonso, S.; Dedon, P.C. Identification of N6,N6-dimethyladenosine in transfer RNA from Mycobacterium bovis Bacille Calmette-Guérin. Molecules 2011, 16, 5168–5181. [Google Scholar] [CrossRef] [PubMed]
- Metodiev, M.D.; Lesko, N.; Park, C.B.; Cámara, Y.; Shi, Y.; Wibom, R.; Hultenby, K.; Gustafsson, C.M.; Larsson, N.G. Methylation of 12S rRNA is necessary for in vivo stability of the small subunit of the mammalian mitochondrial ribosome. Cell Metab. 2009, 9, 386–397. [Google Scholar] [CrossRef] [PubMed]
- Helser, T.L.; Davies, J.E.; Dahlberg, J.E. Change in methylation of 16S ribosomal RNA associated with mutation to Kasugamycin resistance in Escherichia coli. Nature 1971, 233, 12–14. [Google Scholar] [CrossRef]
- Poldermans, B.; Bakker, H.; van Knippenberg, P.H. Studies on the function of two adjacent N6,N6-dimethyladenosines near the 3’ end of 16S ribosomal RNA of Escherichia coli. IV. The effect of the methylgroups on ribosomal subunit interaction. Nucleic Acids Res. 1980, 8, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yue, Y.; Han, D.; Wang, X.; Fu, Y.; Zhang, L.; Jia, G.; Yu, M.; Lu, Z.; Deng, X.; et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014, 10, 93–95. [Google Scholar] [CrossRef] [PubMed]
- Bokar, J.A.; Rath-Shambaugh, M.E.; Ludwiczak, R.; Narayan, P.; Rottman, F. Characterization and partial purification of mRNA N6-adenosine methyltransferase from HeLa cell nuclei. Internal mRNA methylation requires a multisubunit complex. J. Biol. Chem. 1994, 269, 17697–17704. [Google Scholar] [PubMed]
- Bergmann, W.; Feeney, R.J. Contributions to the study of marine products. XXXII. The nucleosides of sponges. I.1. J. Org. Chem. 1951, 16, 981–987. [Google Scholar] [CrossRef]
- Tortora, G.; Ciardiello, F.; Pepe, S.; Tagliaferri, P.; Ruggiero, A.; Bianco, C.; Guarrasi, R.; Miki, K.; Bianco, A. Phase I clinical study with 8-chloro-cAMP and evaluation of immunological effects in cancer patients. Clin. Cancer Res. 1995, 1, 377–384. [Google Scholar] [PubMed]
- Krett, N.L.; Davies, K.M.; Ayres, M.; Ma, C.; Nabhan, C.; Gandhi, V.; Rosen, S.T. 8-amino-adenosine is a potential therapeutic agent for multiple myeloma. Mol. Cancer Ther. 2004, 3, 1411–1420. [Google Scholar] [PubMed]
- Stellrecht, C.M.; Vangapandu, H.V.; Le, X.-F.; Mao, W.; Shentu, S. ATP directed agent, 8-chloro-adenosine, induces AMP activated protein kinase activity, leading to autophagic cell death in breast cancer cells. J. Hematol. Oncol. 2014, 7, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Kearney, A.Y.; Fan, Y.-H.; Giri, U.; Saigal, B.; Gandhi, V.; Heymach, J.V.; Zurita, A.J. 8-chloroadenosine sensitivity in renal cell carcinoma is associated with AMPK activation and mTOR pathway inhibition. PLoS ONE 2015, 10, e0135962. [Google Scholar] [CrossRef] [PubMed]
- Dennison, J.B.; Shanmugam, M.; Ayres, M.L.; Qian, J.; Krett, N.L.; Medeiros, L.J.; Neelapu, S.S.; Rosen, S.T.; Gandhi, V. 8-aminoadenosine inhibits Akt/mTOR and Erk signaling in mantle cell lymphoma. Blood 2010, 116, 5622–5630. [Google Scholar] [CrossRef] [PubMed]




© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vaden, R.M.; Oswald, N.W.; Potts, M.B.; MacMillan, J.B.; White, M.A. FUSION-Guided Hypothesis Development Leads to the Identification of N6,N6-Dimethyladenosine, a Marine-Derived AKT Pathway Inhibitor. Mar. Drugs 2017, 15, 75. https://doi.org/10.3390/md15030075
Vaden RM, Oswald NW, Potts MB, MacMillan JB, White MA. FUSION-Guided Hypothesis Development Leads to the Identification of N6,N6-Dimethyladenosine, a Marine-Derived AKT Pathway Inhibitor. Marine Drugs. 2017; 15(3):75. https://doi.org/10.3390/md15030075
Chicago/Turabian StyleVaden, Rachel M., Nathaniel W. Oswald, Malia B. Potts, John B. MacMillan, and Michael A. White. 2017. "FUSION-Guided Hypothesis Development Leads to the Identification of N6,N6-Dimethyladenosine, a Marine-Derived AKT Pathway Inhibitor" Marine Drugs 15, no. 3: 75. https://doi.org/10.3390/md15030075
APA StyleVaden, R. M., Oswald, N. W., Potts, M. B., MacMillan, J. B., & White, M. A. (2017). FUSION-Guided Hypothesis Development Leads to the Identification of N6,N6-Dimethyladenosine, a Marine-Derived AKT Pathway Inhibitor. Marine Drugs, 15(3), 75. https://doi.org/10.3390/md15030075