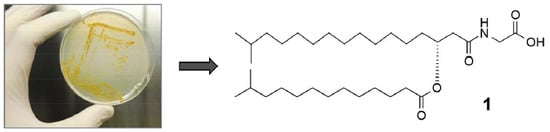
Linear Aminolipids with Moderate Antimicrobial Activity from the Antarctic Gram-Negative Bacterium Aequorivita sp.
Abstract
:1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. General Experimental Procedures
4.2. Isolation and Identification of Biological Material
4.3. Fermentation, Extraction and Isolation
4.4. LC-HRMS2
4.5. Anti-MRSA Assay
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Marcy, Y.; Ouverney, C.; Bik, E.M.; Lösekann, T.; Ivanova, N.; Martin, H.G.; Szeto, E.; Platt, D.; Hugenholtz, P.; Relman, D.A.; et al. Dissecting biological “dark matter” with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth. Proc. Natl. Acad. Sci USA 2007, 104, 11889–11894. [Google Scholar] [CrossRef] [PubMed]
- Kaeberlein, T.; Lewis, K.; Epstein, S.S. Isolating “uncultivable” microorganisms in pure culture in a simulated natural environment. Science 2002, 296, 1127–1129. [Google Scholar] [CrossRef] [PubMed]
- Nichols, D.; Cahoon, N.; Trakhtenberg, E.M.; Pham, L.; Mehta, A.; Belanger, A.; Kanigan, T.; Lewis, K.; Epstein, S.S. Use of ichip for high-throughput in situ cultivation of “uncultivable” microbial species. Appl. Environ. Microbiol. 2010, 76, 2445–2450. [Google Scholar] [CrossRef] [PubMed]
- Ling, L.L.; Schneider, T.; Peoples, A.J.; Spoering, A.L.; Engels, I.; Conlon, B.P.; Mueller, A.; Schäberle, T.F.; Hughes, D.E.; Epstein, S.; et al. A new antibiotic kills pathogens without detectable resistance. Nature 2015, 517, 455–459. [Google Scholar] [CrossRef] [PubMed]
- Ingham, C.J.; Sprenkels, A.; Bomer, J.; Molenaar, D.; van den Berg, A.; van Hylckama Vlieg, J.E.; de Vos, W.M. The micro-Petri dish, a million-well growth chip for the culture and high-throughput screening of microorganisms. Proc. Natl. Acad. Sci. USA 2007, 104, 18217–18222. [Google Scholar] [CrossRef] [PubMed]
- Catón, L.; Yurkov, A.; Giesbers, M.; Dijksterhuis, J.; Ingham, C.J. Physically triggered morphology changes in a novel Acremonium isolate cultivated in precisely engineered microfabricated environments. Front. Microbiol. 2017, 8, 1269–1282. [Google Scholar] [CrossRef] [PubMed]
- Masschelein, J.; Jenner, M.; Challis, G.L. Antibiotics from Gram-negative bacteria: A comprehensive overview and selected biosynthetic highlights. Nat. Prod. Rep. 2017, 34, 712–783. [Google Scholar] [CrossRef] [PubMed]
- Lorig-Roach, N.; Still, P.C.; Coppage, D.; Compton, J.E.; Crews, M.S.; Navarro, G.; Tenney, K.; Crews, P. Evaluating nitrogen-containing biosynthetic products produced by saltwater culturing of several California littoral zone Gram-negative bacteria. J. Nat. Prod. 2017, 80, 2304–2310. [Google Scholar] [CrossRef] [PubMed]
- Tsukimoto, M.; Nagaoka, M.; Shishido, Y.; Fujimoto, J.; Nishisaka, F.; Matsumoto, S.; Harunari, E.; Imada, C.; Matsuzaki, T. Bacterial production of the tunicate-derived antitumor cyclic depsipeptide didemnin B. J. Nat. Prod. 2011, 74, 2329–2331. [Google Scholar] [CrossRef] [PubMed]
- Sudek, S.; Lopanik, N.B.; Waggoner, L.E.; Hildebrand, M.; Anderson, C.; Liu, H.; Patel, A.; Sherman, D.H.; Haygood, M.G. Identification of the putative bryostatin polyketide synthase gene cluster from “Candidatus Endobugula sertula”, the uncultivated microbial symbiont of the marine bryozoan Bugula neritina. J. Nat. Prod. 2007, 70, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Timmermans, M.L.; Paudel, Y.P.; Ross, A.C. Investigating the biosynthesis of natural products from marine Proteobacteria: A survey of molecules and strategies. Mar. Drugs. 2017, 15, 235–271. [Google Scholar] [CrossRef] [PubMed]
- Still, P.C.; Johnson, T.A.; Theodore, C.M.; Loveridge, S.T.; Crews, P. Scrutinizing the scaffolds of marine biosynthetics from different source organisms: Gram-negative cultured bacterial products enter center stage. J. Nat. Prod. 2014, 77, 690–702. [Google Scholar] [CrossRef] [PubMed]
- Bowman, J.P.; Nichols, D.S. Aequorivita gen. nov., a member of the family Flavobacteriaceae isolated from terrestrial and marine Antarctic habitats. Int. J. Syst. Evol. Microbiol. 2002, 52, 1533–1541. [Google Scholar] [PubMed]
- Kawazowe, R.; Okuyama, H.; Reichardt, W.; Sasaki, S. Phospholipids and a novel glycine-containing lipoamino acid in Cytophaga johnsonae Stanier strain C21. J. Bacteriol. 1991, 173, 5470–5475. [Google Scholar] [CrossRef]
- Morishita, T.; Sato, A.; Hisamoto, M.; Oda, T.; Matsuda, K.; Ishii, A.; Kodama, K. N-Type calcium channel blockers from a marine bacterium, Cytophaga sp. SANK71996. J. Antibiot. 1997, 50, 457–468. [Google Scholar] [CrossRef] [PubMed]
- Batrakov, S.G.; Nikitin, D.I.; Mosezhnyi, A.E.; Ruzhitsky, A.O. A glycine-containing phosphorus-free lipoaminoacid from the gram-negative marine bacterium Cyclobacterium marinus WH. Chem. Phys. Lipids 1999, 99, 139–143. [Google Scholar] [CrossRef]
- Yoshida, K.; Iwami, M.; Umehara, Y.; Nishikawa, M.; Uchida, I.; Kohsaka, M.; Aoki, H.; Imanaka, H. Studies on WB-3559 A, B, C and D, new potent fibrinolytic agents I. Discovery, identification, isolation and characterization. J. Antibiot. 1985, 38, 1469–1475. [Google Scholar] [CrossRef] [PubMed]
- Uchida, I.; Yoshida, C.K.; Kawai, Y.; Takase, S.; Itoh, Y.; Tanaka, H.; Kohsaka, M.; Imanaka, H. Studies on WB-3559 A, B, C and D, new potent fibrinolytic agents II. Structure and synthesis. J. Antibiot. 1985, 38, 1476–1486. [Google Scholar] [CrossRef] [PubMed]
- Morii, H.; Nishihara, M.; Ohga, M.; Koga, Y. A diphytanyl ether analog of phosphatidyl serine from a methanogenic bacterium, Methanobacterium arboriphilus . J. Lipid Res. 1986, 27, 724–730. [Google Scholar]
- Shahina, M.; Hameed, A.; Lin, S.Y.; Lai, W.A.; Liu, Y.C.; Hsu, Y.H.; Young, C.C. Luteibaculum oceani gen. nov., sp. nov., a carotenoid-producing, lipolytic bacterium isolated from surface seawater, and emended description of the genus Owenweeksia Lau et al. 2005. Int. J. Syst. Evol. Microbiol. 2013, 63, 4765–4770. [Google Scholar] [CrossRef] [PubMed]
- Nedashkovskaya, O.I.; Kim, S.G.; Zhukova, N.V.; Mikhailov, V.V. Olleya algicola sp. nov., a marine bacterium isolated from the green alga Ulva fenestrata. Int. J. Syst. Evol. Microbiol. 2017, 67, 2205–2210. [Google Scholar] [CrossRef] [PubMed]
- Baek, K.; Lee, Y.M.; Hwang, C.Y.; Park, H.; Jung, Y.J.; Kim, M.K.; Hong, S.G.; Kim, J.H.; Lee, H.K. Psychroserpens jangbogonensis sp. nov., a psychrophilic bacterium isolated from Antarctic marine sediment. Int. J. Syst. Evol. Microbiol. 2015, 65, 183–188. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Jiang, F.; Chang, X.; Qiu, X.; Ren, L.; Qu, Z.; Deng, S.; Da, X.; Fang, C.; Peng, F. Flavobacterium collinsense sp. nov., isolated from a till sample of an Antarctic glacier. Int. J. Syst. Evol. Microbiol. 2016, 66, 172–177. [Google Scholar] [CrossRef] [PubMed]
- Tahara, Y.; Kameda, M.; Yamada, M.; Kondo, K. A new lipid; the ornithine and taurine-containing “cerilipin”. Agric. Biol. Chem. 1976, 40, 243–244. [Google Scholar]
- Kawai, Y.; Yano, I.; Kaneda, K. Various kinds of lipoamino acids including a novel serine-containing lipid in an opportunistic pathogen Flavobacterium. Their structures and biological activities on erythrocytes. Eur. J. Biochem. 1988, 171, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Kawai, Y.; Akagawa, K. Macrophage activation of an ornithine-containing lipid or a serine-containing lipid. Infect. Immun. 1989, 57, 2086–2091. [Google Scholar] [PubMed]
- Tahara, Y.; Yamada, Y.; Kondo, K. A New lysine-containing lipid isolated from Agrobacterium tumefaciens. Agric. Biol. Chem. 1976, 40, 1449–1450. [Google Scholar] [CrossRef]
- Thasana, N.; Prapagdee, B.; Rangkadilok, N.; Sallabhan, R.; Aye, S.L.; Ruchirawat, S.; Loprasert, S. Bacillus subtilis SSE4 produces subtulene A, a new lipopeptide antibiotic possessing an unusual C15 unsaturated beta-amino acid. FEBS Lett. 2010, 584, 3209–3214. [Google Scholar] [CrossRef] [PubMed]
- Sivasamy, A.; Krishnaveni, M.; Rao, P.G. Preparation, characterization, and surface and biological properties of N-stearoyl amino acids. JAOCS 2001, 78, 897–902. [Google Scholar] [CrossRef]
- Rezanka, T.; Sigler, K. Odd-numbered very-long-chain fatty acids from the microbial, animal and plant kingdoms. Prog. Lipid Res. 2009, 48, 206–238. [Google Scholar] [CrossRef] [PubMed]


| Position | 1 | 2 | 3 |
|---|---|---|---|
| δH, Mult. (J in Hz) | δH, Mult. (J in Hz) | δH, Mult. (J in Hz) | |
| 2 | 2.54, m | 2.51, m | 2.50, m |
| 3 | 5.16, m | 5.17, m | 5.17, m |
| 4 | 1.62 a | 1.60, m | 1.60, m |
| 5 | 1.30 a | 1.30 a | 1.30 a |
| 6–13 | 1.25 a | 1.25 a | 1.25 a |
| 14 | 1.14, m | 1.14, m | 1.14, m |
| 15 | 1.51, m | 1.51, m | 1.51, m |
| 16 | 0.86, d (6.7) | 0.86, d (6.7) | 0.86, d (6.7) |
| 17 | 0.86, d (6.7) | 0.86, d (6.7) | 0.86, d (6.7) |
| 2′ | 2.30, t (7.5) | 2.34 a | 2.37 a |
| 3′ | 1.60 a | 2.34 a | 2.36 a |
| 4′ | 1.28 a | 5.30, m | 5.30, m |
| 5′ | 1.25 a | 5.40, m | 5.42, m |
| 6′ | 1.25 a | 2.02, m | 2.04, m |
| 7′ | 1.25 a | 1.33, m | 1.33, m |
| 8′–10′ | 1.25 a | 1.25, m | 1.25, m |
| 11′ | 1.14, m | 1.25 a | 1.25 a |
| 12′ | 1.51, m | 1.14, m | 1.14, m |
| 13′ | 0.86, d (6.7) | 1.51, m | 1.51, m |
| 14′ | 0.86, d (6.7) | 0.86, d (6.7) | 0.86, d (6.7) |
| 15′ | 0.86, d (6.7) | 0.86, d (6.7) | |
| 2′′ | 4.07, d (5.0) | 4.03, d (5.3) | 4.03, d (5.3) |
| 2′′′ | 4.66, m | ||
| 3′′′ | 4.00 a 3.96 a | ||
| OCH3 | 3.75, s | 3.75, s | |
| (Gly)NH | 6.38, brt (5.3) | 6.25, brt (5.0) | 6.38, t (4.5) |
| (Ser)NH | 6.90, d (7.7) |
| Position | 1 a | 2 | 3 |
|---|---|---|---|
| δC, Type | δC, Type | δC, Type | |
| 1 | 170.5, C | 169.9, C | 170.8, C |
| 2 | 41.4, CH2 | 41.2, CH2 | 41.7, CH2 |
| 3 | 71.1, CH | 71.3, CH | 71.5, CH |
| 4 | 34.1, CH2 | 34.3, CH2 | 34.2, CH2 |
| 5 | 25.4, CH2 | 29.5, CH2 | 29.6, CH2 |
| 6–13 | 29.0, CH2 | 29.0, CH2 | 29.0, CH2 |
| 14 | 39.1, CH2 | 39.1, CH2 | 39.1, CH2 |
| 15 | 27.8, CH | 27.8, CH | 27.8, CH |
| 16 | 22.7, CH3 | 22.7, CH3 | 22.7, CH3 |
| 17 | 22.7, CH3 | 22.7, CH3 | 22.7, CH3 |
| 1′ | 173.7, C | 173.9, C | 173.7, C |
| 2′ | 34.5, CH2 | 34.4, CH2 | 34.4, CH2 |
| 3′ | 24.9, CH2 | 22.7, CH2 | 22.6, CH2 |
| 4′ | 29.0, CH2 | 127.2, CH | 127.1, CH |
| 5′ | 29.0, CH2 | 131.6, CH | 131.7, CH |
| 6′ | 29.0, CH2 | 27.1, CH2 | 27.1, CH2 |
| 7′ | 29.0, CH2 | 29.6, CH2 | 29.5, CH2 |
| 8′–10′ | 29.0, CH2 | 29.0, CH2 | 29.0, CH2 |
| 11′ | 39.1, CH2 | 29.0, CH2 | 29.0, CH2 |
| 12′ | 27.8, CH | 39.1, CH2 | 39.1, CH2 |
| 13′ | 22.7, CH3 | 27.8, CH | 27.8, CH |
| 14′ | 22.7, CH3 | 22.7, CH3 | 22.7, CH3 |
| 15′ | 22.7, CH3 | 22.7, CH3 | |
| 1′′ | 171.1, C | 170.2, C | 170.0, C |
| 2′′ | 41.1, CH2 | 41.2, CH2 | 41.1, CH2 |
| 1′′′ | 170.6, C | ||
| 2′′′ | 54.8, CH2 | ||
| 3′′′ | 62.7, CH | ||
| OCH3 | 52.0, CH3 | 52.0, CH3 |
| Sample | IC50 Value (µg/mL) |
|---|---|
| EtOAc extract | 120 |
| 1 | 58 |
| 2 | 145 |
| 3 | >200 |
| 4 | 22 |
| 5 | 93 |
| 6 | >200 |
| 7 | >200 |
| Reference drug | 2.89 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chianese, G.; Esposito, F.P.; Parrot, D.; Ingham, C.; De Pascale, D.; Tasdemir, D. Linear Aminolipids with Moderate Antimicrobial Activity from the Antarctic Gram-Negative Bacterium Aequorivita sp. Mar. Drugs 2018, 16, 187. https://doi.org/10.3390/md16060187
Chianese G, Esposito FP, Parrot D, Ingham C, De Pascale D, Tasdemir D. Linear Aminolipids with Moderate Antimicrobial Activity from the Antarctic Gram-Negative Bacterium Aequorivita sp. Marine Drugs. 2018; 16(6):187. https://doi.org/10.3390/md16060187
Chicago/Turabian StyleChianese, Giuseppina, Fortunato Palma Esposito, Delphine Parrot, Colin Ingham, Donatella De Pascale, and Deniz Tasdemir. 2018. "Linear Aminolipids with Moderate Antimicrobial Activity from the Antarctic Gram-Negative Bacterium Aequorivita sp." Marine Drugs 16, no. 6: 187. https://doi.org/10.3390/md16060187
APA StyleChianese, G., Esposito, F. P., Parrot, D., Ingham, C., De Pascale, D., & Tasdemir, D. (2018). Linear Aminolipids with Moderate Antimicrobial Activity from the Antarctic Gram-Negative Bacterium Aequorivita sp. Marine Drugs, 16(6), 187. https://doi.org/10.3390/md16060187