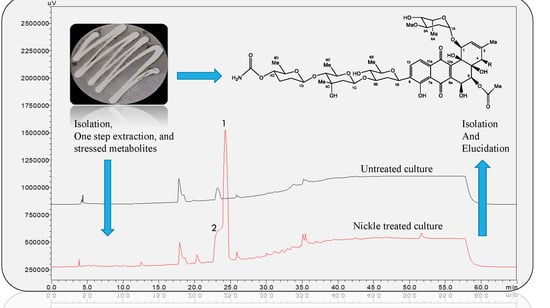
Stress-Driven Discovery of New Angucycline-Type Antibiotics from a Marine Streptomyces pratensis NA-ZhouS1
Abstract
:1. Introduction
2. Results
Structural Elucidation of Novel Compounds
3. Discussion
4. Materials and Methods
4.1. General Experimental Procedures
4.2. Isolation and Identification of Streptomyces sp. NA-ZhouS1
4.3. Analysis of Normal Culture and Metal Stress Cultivation
4.4. Large Scale Fermentation, Extraction and Isolation
4.5. Antimicrobial Activity of Stressed Metabolites
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Magdy, W.; Abdel-Motaal, F.F.; El-Zayat, S.A.; El-Sayed, M.A.E.; Helaly, S.E. Nigragillin, Nigerazine B and five Naphtho-γ-pyrones from Aspergillus japonicus isolated from hot desert soil. Nat. Prod. J. 2017, 7, 216–223. [Google Scholar] [CrossRef]
- Ahmad, N.; Fatma, T. Production of indole-3-acetic acid by cyanobacterial strains. Nat. Prod. J. 2017, 7, 112–120. [Google Scholar] [CrossRef]
- Gupta, A.; Singh, S. Characterization of NaCl-tolerant mutant strain of the cyanobacterium Spirulina platensis overproducing phycocyanin. Nat. Prod. J. 2017, 7, 153–164. [Google Scholar] [CrossRef]
- Shi, Y.; Pan, C.; Auckloo, B.N.; Chen, X.; Chen, C.T.A.; Wang, K.; Wu, X.; Ye, Y.; Wu, B. Stress-driven discovery of a cryptic antibiotic produced by Streptomyces sp. WU20 from Kueishantao hydrothermal vent with an integrated metabolomics strategy. Appl. Microbiol. Biotechnol. 2017, 101, 1395–1408. [Google Scholar] [CrossRef] [PubMed]
- Montaser, R.; Luesch, H. Marine natural products: A new wave of drugs? Future Med. Chem. 2012, 3, 1475–1489. [Google Scholar] [CrossRef] [PubMed]
- Blunt, J.W.; Copp, B.R.; Keyzers, R.A.; Munro, M.H.G.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2017, 34, 235–294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdelfattah, M.S.; Kharel, M.K.; Hitron, J.A.; Baig, I. Moromycins A and B, Isolation and structure elucidation of C-glycosylangucycline-type antibiotics from Streptomyces sp. KY002. Nat. Prod. J. 2008, 10, 1569–1573. [Google Scholar] [CrossRef] [PubMed]
- Rao, M.; Wei, W.; Ge, M.; Chen, D.; Sheng, X. A new antibacterial lipopeptide found by UPLC-MS from an actinomycete Streptomyces sp. HCCB10043. Nat. Prod. Res. 2013, 27, 2190–2195. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Yang, X.; Zhang, Y.; Zhou, H.; Zhang, J.; Xu, L.; Ding, Z. A new daidzein derivative from endophytic Streptomyces sp. YIM 65408. Nat. Prod. Res. 2013, 27, 1727–1731. [Google Scholar] [CrossRef] [PubMed]
- Awad, H.M.; El-Enshasy, H.A.; Hanapi, S.Z.; Hamed, E.R.; Rosidi, B. A new chitinase-producer strain Streptomyces glauciniger WICC-A03: Isolation and identification as a biocontrol agent for plants phytopathogenic fungi. Nat. Prod. Res. 2014, 28, 2273–2277. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Sun, W.; Yu, Y.; Li, Z.; Deng, Z.; Lin, S. A new glutarimide derivative from marine sponge-derived Streptomyces anulatus S71. Nat. Prod. Res. 2014, 28, 1602–1606. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Zhang, Y.; Yin, X.; Wang, S. Nesterenkonia alkaliphila sp. Nov., an alkaliphilic, halotolerant actinobacteria isolated from the western pacific ocean. Int. J. Syst. Evol. Microbiol. 2015, 65, 516–521. [Google Scholar] [CrossRef] [PubMed]
- Balachandran, C.; Duraipandiyan, V.; Emi, N.; Ignacimuthu, S. Antimicrobial and cytotoxic properties of Streptomyces sp. (ERINLG-51) isolated from Southern Western Ghats. South Indian J. Biol. Sci. 2015, 1, 7–14. [Google Scholar] [CrossRef]
- Hu, Z.; Qin, L.; Wang, Q.; Ding, W.; Chen, Z.; Ma, Z. Angucycline antibiotics and its derivatives from marine-derived actinomycete Streptomyces sp. A6H. Nat. Prod. Res. 2016, 30, 2551–2558. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Zhang, L.; Guo, X.; Dai, X.; Liu, L.; Xi, L.; Wang, J.; Song, L.; Wang, Y.; Zhu, Y.; et al. Diversity, biogeography, and biodegradation potential of actinobacteria in the deep-sea sediments along the southwest Indian ridge. Front. Microbiol. 2016, 7, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Helaly, S.E.; Goodfellow, M.; Zinecker, H.; Imhoff, J.F.; Süssmuth, R.D.; Fiedler, H.P. Warkmycin, a novel angucycline antibiotic produced by Streptomyces sp. Acta 2930. J. Antibiot. (Tokyo) 2013, 66, 669–674. [Google Scholar] [CrossRef] [PubMed]
- Ma, M.; Rateb, M.E.; Teng, Q.; Yang, D.; Rudolf, J.D.; Zhu, X.; Huang, Y.; Zhao, L.X.; Jiang, Y.; Li, X.; et al. Angucyclines and Angucyclinones from Streptomyces sp. CB01913 featuring C-ring cleavage and expansion. J. Nat. Prod. 2015, 78, 2471–2480. [Google Scholar] [CrossRef] [PubMed]
- Zotchev, S.B. Marine actinomycetes as an emerging resource for the drug development pipelines. J. Biotechnol. 2012, 158, 168–175. [Google Scholar] [CrossRef] [PubMed]
- Feling, R.H.; Buchanan, G.O.; Mincer, T.J.; Kauffman, C.A.; Jensen, P.R.; Fenical, W. Salinosporamide A: A highly cytotoxic proteasome inhibitor from a novel microbial source, a marine bacterium of the new genus Salinospora. Angew. Chem. Int. Ed. 2003, 42, 355–357. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Jiang, W.; Auckloo, B.N.; Wu, B. Several classes of natural products with metal ion chelating ability. Curr. Org. Chem. 2015, 19, 1935–1953. [Google Scholar] [CrossRef]
- Auckloo, B.N.; Pan, C.; Akhter, N.; Wu, B.; Wu, X.; He, S. Stress-driven discovery of novel cryptic antibiotics from a marine fungus Penicillium sp. BB1122. Front. Microbiol. 2017, 8, 1450. [Google Scholar] [CrossRef] [PubMed]
- Ding, C.; Wu, X.; Auckloo, B.N.; Chen, C.T.A.; Ye, Y.; Wang, K.; Wu, B. An unusual stress metabolite from a hydrothermal vent fungus Aspergillus sp. WU 243 induced by cobalt. Molecules 2016, 21, 105. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Zhong, Y.; Shen, L.; Wu, X.; Ye, Y.; Chen, C.-T.; Wu, B. Stress-driven discovery of natural products from extreme marine environment- Kueishantao hydrothermal vent, a case study of metal switch valve. Curr. Org. Chem. 2014, 18, 925–934. [Google Scholar] [CrossRef]
- Shaaban, K.A.; Ahmed, T.A.; Leggas, M.; Rohr, J. Saquayamycins G–K, cytotoxic angucyclines from Streptomyces sp. including two analogues bearing the Amino-sugar Rednose. J. Nat. Prod. 2012, 75, 1383–1392. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Xu, Q.; Bu, Q.; Guo, Y.; Liu, S.; Liu, Y.; Du, Y.; Li, Y. Genome mining-directed activation of a silent angucycline biosynthetic gene cluster in Streptomyces chattanoogensis. Chembiochem 2015, 16, 496–502. [Google Scholar] [CrossRef] [PubMed]
- Rohr, J.; Thiericke, R. Angucycline group antibiotics. Nat. Prod. Rep. 1992, 9, 103–137. [Google Scholar] [CrossRef] [PubMed]
- Kharel, M.K.; Pahari, P.; Shepherd, M.D.; Tibrewal, N.; Nybo, S.E.; Shaaban, K.A.; Rohr, J. Angucyclines: Biosynthesis, mode-of-action, new natural products, and synthesis. Nat. Prod. Rep. 2012, 29, 264–325. [Google Scholar] [CrossRef] [PubMed]
- Uesato, S.; Tokunaga, T.; Mizuno, Y.; Fujioka, H.; Kada, S.; Kuwajima, H. Absolute stereochemistry of gastric antisecretory compound P371A1 and its congener P371A2 from Streptomyces species P371. J. Nat. Prod. 2000, 63, 787–792. [Google Scholar] [CrossRef] [PubMed]
- Xue, Z.R.; Xu, W.P.; Chen, J.; Tao, L.M. Study of Streptomyces griseolus CGMCC 1370 and its herbicidal metabolite. Chin. J. Pestic. Sci. 2011, 13, 155–161. [Google Scholar]
- Cronan, J.M.; Davidson, T.R.; Singleton, F.L.; Colwell, R.R.; Cardellina, J.H. Plant growth promoters isolated from a marine bacterium associated with Palythoa sp. Nat. Prod. Lett. 1998, 11, 271–278. [Google Scholar] [CrossRef]
- Maskey, R.P.; Shaaban, M.; Gru, I. Quinazolin-4-one derivatives from Streptomyces isolates. Nat. Prod. J. 2004, 7, 1131–1134. [Google Scholar] [CrossRef] [PubMed]
- Sharipov, B.T.; Pilipenko, A.N.; Valeev, F.A. Eleuthesides and their analogs: VIII. Preparation of menthane derivatives from levoglucosenone and (2E,4E)-6-methylhepta-2,4-dienyl acetate by Diels-Alder reaction. Russ. J. Org. Chem. 2014, 50, 1628–1635. [Google Scholar] [CrossRef]
- Schneemann, I.; Kajahn, I.; Ohlendorf, B.; Zinecker, H.; Erhard, A.; Nagel, K.; Wiese, J.; Imhoff, J.F. Mayamycin, a cytotoxic polyketide from a Streptomyces strain isolated from the marine sponge Halichondria panicea. J. Nat. Prod. 2010, 73, 1309–1312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdelfattah, M.; Maskey, R.P.; Asolkar, R.N.; Grün-Wollny, I.; Laatsch, H. Seitomycin: Isolation, structure elucidation and biological activity of a new angucycline antibiotic from a terrestrial Streptomycete. J. Antibiot. (Tokyo) 2003, 56, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.B.; Xu, W.P.; Ye, G.Y.; Zhang, Y.L.; Ni, W.W.; Tao, L.M. The herbicidal active component derived from soil actinomycete SPRI 70014. Chin. J. Antibiot. 2008, 33, 461–466. [Google Scholar]
- Morishita, T.; Sato, A.; Ando, T.; Oizumi, K.; Miyamoto, M.; Enokita, R.; OKAZAKI, T. A novel bone resorption inhibitor, A-75943 isolated from Streptomyces sp. SANK 61296. J. Antibiot. (Tokyo) 1998, 51, 531–538. [Google Scholar] [CrossRef] [PubMed]
- Kalinovskaya, N.I.; Kalinovsky, A.I.; Romanenko, L.A.; Pushilin, M.A.; Dmitrenok, P.S.; Kuznetsova, T.A. New angucyclinones from the marine mollusk associated actinomycete Saccharothrix espanaensis An 113. Nat. Prod. Commun. 2008, 3, 1611–1616. [Google Scholar]
- Fotso, S.; Mahmud, T.; Zabriskie, T.M.; Santosa, D.A.; Proteau, P.J. Angucyclinones from an Indonesian Streptomyces sp. J. Nat. Prod. 2007, 71, 61–65. [Google Scholar] [CrossRef] [PubMed]
- Fedoryshyn, M.; Nur-e-Alam, M.; Zhu, L.; Luzhetskyy, A.; Rohr, J.; Bechthold, A. Surprising production of a new urdamycin derivative by S. fradiae∆urdQ/R. J. Biotechnol. 2007, 130, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Pan, G.; Zhu, X.; Fan, K.; Gao, W.; Ai, G.; Ren, J.; Shi, M.; Olano, C.; Salas, J.A. Engineered jadomycin analogues with altered sugar moieties revealing JadS as a substrate flexible O-glycosyltransferase. Appl. Microbiol. Biotechnol. 2017, 13, 5291–5300. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Yang, T.; Ren, X.; Liu, J.; Song, Y.; Sun, A.; Ma, J.; Wang, B.; Zhang, Y.; Huang, C. Cytotoxic angucycline class glycosides from the deep sea actinomycete Streptomyces lusitanus SCSIO LR32. J. Nat. Prod. 2012, 75, 202–208. [Google Scholar] [CrossRef] [PubMed]
- Henkel, T.; Rohr, J.; Beale, J.M.; Schwenen, L. Landomycins, new angucycline antibiotics from Streptomyces sp. J. Antibiot. (Tokyo) 1990, 43, 492–503. [Google Scholar] [CrossRef] [PubMed]
- Rong, X.; Doroghazi, J.R.; Cheng, K.; Zhang, L.; Buckley, D.H.; Huang, Y. Classification of Streptomyces phylogroup pratensis (Doroghazi and Buckley, 2010) based on genetic and phenotypic evidence, and proposal of Streptomyces pratensis sp. nov. Syst. Appl. Microbiol. 2013, 36, 401–407. [Google Scholar] [CrossRef] [PubMed]
- Doroghazi, J.R.; Buckley, D.H. Intraspecies comparison of Streptomyces pratensis genomes reveals high levels of recombination and gene conservation between strains of disparate geographic origin. BMC Genomics 2014, 15, 1–14. [Google Scholar] [CrossRef] [PubMed]




| Position | δC, Type | δH (Mult., J in Hz) | HMBC | COSY |
|---|---|---|---|---|
| 1 | 82.1, CH | 4.34, d (4.2) | C-1A, C-2, C-12b, C-4a | H-2 |
| 2 | 120.8, CH | 5.63, s | C-3 | H-1 |
| 3 | 136.6, C | - | - | - |
| 4 | 36.3, CH2 | 1.99, d (17.6); 2.36, d (17.6) | Me-3, C-4a | - |
| 4a | 75.6, C | - | --- | - |
| 5 | 76.2, CH | 5.84, d, (6.8) | C-6, COMe | H-6 |
| 6 | 69.9, CH | 4.92, d, (6.8) | C-5, C-6a, C-12a | H-5 |
| 6a | 145.2, C | - | - | - |
| 7 | 190.5, C | - | - | - |
| 7a | 115.6, C | - | - | - |
| 8 | 158.6, C | - | - | - |
| 9 | 139.3, C | - | - | - |
| 10 | 134.2, CH | 7.87, d (7.9) | C-7a, C-8, C-1B, C11a | H-11 |
| 11 | 120.1, CH | 7.64, d (7.9) | C-7a, C-9, C-12 | H-10 |
| 11a | 132.6, C | - | - | - |
| 12 | 187.2, C | - | - | - |
| 12a | 146.0, C | - | - | - |
| 12b | 78.5, C | - | - | - |
| 3-Me | 23.5, CH3 | 1.68, s | C-3, C-4 | - |
| 5-COMe | 172.3, C | - | - | - |
| 20.9, CH3 | 2.20, s | - | - | |
| Sugar A | ||||
| 1A | 100.6, CH | 4.60, d (4.0) | C-1, C-3A, C-5A | H-2A |
| 2A | 30.6, CH2 | 1.31, overlapped; 1.88, m | - | H-1A, H-3A |
| 3A | 78.4, CH | 3.29, m | OMe-3A | H-4A |
| 4A | 66.9, CH | 3.49, m | - | H-5A |
| 5A | 63.4, CH | 4.29, m | - | H-6A |
| 6A | 16.8, CH3 | 1.18, d (6.6) | C-5A | H-5A |
| OMe-3A | 57.5,C | 3.24, s | - | - |
| Sugar B | ||||
| 1B | 72.4, CH | 4.89, s | C-10, C-9, C-8, C7a, C-2B, C-3B, C-4B, C-5B | H-2B |
| 2B | 38.6, CH2 | 1.42, d (12.6);2.50, dd (12.1, 4.2) | - | H-1B, H-3B |
| 3B | 82.5, CH | 3.84, m | C-1C | H-4B |
| 4B | 76.8, CH | 3.11, t (8.9) | - | H-5B |
| 5B | 77.7, CH | 3.46, m | C-6B | H-6B |
| 6B | 18.8, CH3 | 1.38, d (6.1) | - | - |
| Sugar C | ||||
| 1C | 99.3, CH | 4.78, dd (9.9, 1.8) | C-3B, C-2C | H-2C |
| 2C | 45.6, CH2 | 1.68, overlapped; 1.95, m | C-3C, C-4C | H-1C |
| 3C | 71.5, C | - | - | - |
| 4C | 90.5, CH | 3.19, d (9.6) | C-5C, C-6C, C-1D | H-5C |
| 5C | 71.9, CH | 3.55, m | - | H-6C |
| 6C | 18.5, CH3 | 1.31, d (6.1) | C-5C | H-5C |
| Me-3C | 22.6, CH3 | 1.25, s | C-3C, C-4C | - |
| Sugar D | ||||
| 1D | 104.5, CH | 4.62, s | C-4C, C-2D, C-3D | H-2D |
| 2D | 31.4, CH2 | 1.64, m; 1.99, overlapped | - | H-3D |
| 3D | 28.9, CH2 | 1.57, m; 2.14, m | - | H-4D |
| 4D | 73.9, CH | 4.24, dd (10.0, 4.4) | C-6D, CONH2 | H-5D |
| 5D | 75.3, CH | 3.65, m | - | H-6D |
| 6D | 18.2, CH3 | 1.22, d (6.2) | C-5D | - |
| 4D-CONH2 | 159.6, C | - | C-4D | - |
| Position | δC, Type | δH (Mult., J in Hz) | HMBC | COSY |
|---|---|---|---|---|
| 1 | 81.8, CH | 4.43, d (3.8) | C-1A, C-2, C-3, C-12b, C-4a | H-2 |
| 2 | 123.1, CH | 5.72, d (3.2) | 3-Me, C-4 | H-1 |
| 3 | 138.7, C | - | - | - |
| 4 | 70.5, CH | 4.17, s | 3-Me, C-3, C-4a, C-12b | - |
| 4a | 76.2, C | - | - | - |
| 5 | 75.9, CH | 5.76, d (6.8) | C-6, COMe | H-6 |
| 6 | 69.8, CH | 4.91, m (6.8) | C-5, C-6a, C-12a | H-5 |
| 6a | 144.5, C | - | - | - |
| 7 | 190.8, C | - | - | - |
| 7a | 115.6, C | - | - | - |
| 8 | 158.7, C | - | - | - |
| 9 | 139.4, C | - | - | - |
| 10 | 134.2, CH | 7.85 d (7.7) | C-8, C-1B, C11a | H-11 |
| 11 | 120.0, CH | 7.61, d (7.7) | C-7a, C-9, C-12 | H-10 |
| 11a | 132.5, C | - | - | - |
| 12 | 187.3, C | - | - | - |
| 12a | 146.7, C | - | - | - |
| 12b | 79.7, C | - | - | - |
| 3-Me | 21.9, CH3 | 1.95, s | C-3, C-4 | - |
| 5-COMe | 173.0, C | - | - | - |
| 20.6, CH3 | 2.02, s | - | - | |
| Sugar A | ||||
| 1A | 100.9, CH | 4.57, d (4.4) | C-1, C-3A, C-5A | H-2A |
| 2A | 30.3, CH2 | 1.32, overlapped; 1.85, m | C-1A | H-1A, H-3A |
| 3A | 78.4, CH | 3.27, m | C-4A | H-4A |
| 4A | 67.5, CH | 3.47, m | - | H-5A |
| 5A | 63.7, CH | 4.10, d (6.5) | - | H-6A |
| 6A | 16.8, CH3 | 1.17, d (6.6) | C-5A | H-5A |
| OMe-3A | 57.7, C | 3.30, s | - | - |
| Sugar B | ||||
| 1B | 72.4, CH | 4.87, m (overlapped) | C-9, C-10 | H-2B |
| 2B | 38.6, CH2 | 1.42, m; 2.49, dd (12.3, 4.6) | C-3B, C-4B | H-1B, H-3B |
| 3B | 82.3, CH | 3.84, m | C-4B, C-1C | H-4B |
| 4B | 76.9, CH | 3.11, dd (11.1, 6.7) | C-3B, C-5B, C-6B | H-5B |
| 5B | 77.7, CH | 3.44, dd (6.9, 3.5) | C-6B | H-6B |
| 6B | 18.8, CH3 | 1.37, d (6.1) | C-5B | - |
| Sugar C | ||||
| 1C | 99.3, CH | 4.77, d (9.9) | C-3B, C-2C | H-2C |
| 2C | 45.6, CH2 | 1.95, overlapped; 1.67, m | C-3C, C-4C | H-1C |
| 3C | 71.5, C | - | - | - |
| 4C | 90.5, CH | 3.18, d (9.6) | Me-3C, C-5C, C-6C, C-1D | H-5C |
| 5C | 72.0, CH | 4.81, d (6.5) | --- | H-6C |
| 6C | 18.5, CH3 | 1.30, d (6.1) | C-5C | H-5C |
| Me-3C | 22.6, CH3 | 1.25, s | C-3C, C-4C | - |
| Sugar D | ||||
| 1D | 104.5, CH | 4.61, d (9.2) | C-4C, C-2D, C-3D | H-2D |
| 2D | 31.4, CH2 | 1.99, m; 1.63, m | - | H-3D |
| 3D | 28.9, CH2 | 2.14, m; 1.59, m | - | H-4D |
| 4D | 73.9, CH | 4.24, dd (9.7, 5.5) | C-6D, CONH2 | H-5D |
| 5D | 75.3, CH | 3.62, m | - | H-6D |
| 6D | 18.2, CH3 | 1.22, d (6.1) | C-5D | - |
| 4D-CONH2 | 159.6, C | - | C-4D | - |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akhter, N.; Liu, Y.; Auckloo, B.N.; Shi, Y.; Wang, K.; Chen, J.; Wu, X.; Wu, B. Stress-Driven Discovery of New Angucycline-Type Antibiotics from a Marine Streptomyces pratensis NA-ZhouS1. Mar. Drugs 2018, 16, 331. https://doi.org/10.3390/md16090331
Akhter N, Liu Y, Auckloo BN, Shi Y, Wang K, Chen J, Wu X, Wu B. Stress-Driven Discovery of New Angucycline-Type Antibiotics from a Marine Streptomyces pratensis NA-ZhouS1. Marine Drugs. 2018; 16(9):331. https://doi.org/10.3390/md16090331
Chicago/Turabian StyleAkhter, Najeeb, Yaqin Liu, Bibi Nazia Auckloo, Yutong Shi, Kuiwu Wang, Juanjuan Chen, Xiaodan Wu, and Bin Wu. 2018. "Stress-Driven Discovery of New Angucycline-Type Antibiotics from a Marine Streptomyces pratensis NA-ZhouS1" Marine Drugs 16, no. 9: 331. https://doi.org/10.3390/md16090331
APA StyleAkhter, N., Liu, Y., Auckloo, B. N., Shi, Y., Wang, K., Chen, J., Wu, X., & Wu, B. (2018). Stress-Driven Discovery of New Angucycline-Type Antibiotics from a Marine Streptomyces pratensis NA-ZhouS1. Marine Drugs, 16(9), 331. https://doi.org/10.3390/md16090331