Genome Sequencing of Streptomyces olivaceus SCSIO T05 and Activated Production of Lobophorin CR4 via Metabolic Engineering and Genome Mining
Abstract
:1. Introduction
2. Results and Discussion
2.1. Genome Sequencing and Annotation of Streptomyces olivaceus SCSIO T05
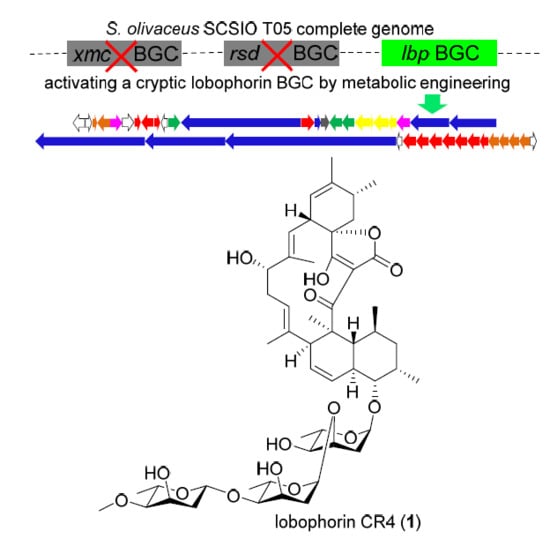
2.2. Activation of a Cryptic Lobophorin BGC in the Genetically Engineered Mutant
2.3. Identification of a Putative Lobophorin (lbp) BGC via Genome Mining
3. Experimental Section
3.1. General Experimental Procedures
3.2. Genome Sequencing and Bioinformatic Analysis
3.3. Construction of a “Triple-Deletion” Mutant Strain
3.4. Fermentation and HPLC-based Analyses of S. olivaceus SCSIO T05 and Its Mutants
3.5. Production, Isolation, and Structure Elucidation of Lobophorin CR4
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Berdy, J. Bioactive microbial metabolites. J. Antibiot. 2005, 58, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2019, 36, 122–173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saha, S.; Zhang, W.; Zhang, G.; Zhu, Y.; Chen, Y.; Liu, W.; Yuan, C.; Zhang, Q.; Zhang, H.; Zhang, L.; et al. Activation and characterization of a cryptic gene cluster reveals a cyclization cascade for polycyclic tetramate macrolactams. Chem. Sci. 2017, 8, 1607–1612. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Zhang, Q.; Tan, B.; Zheng, L.; Li, H.; Zhu, Y.; Zhang, C. Genome Mining and Activation of a Silent PKS/NRPS Gene Cluster Direct the Production of Totopotensamides. Org. Lett. 2017, 19, 5697–5700. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, C.; Liu, C.; Ju, J.; Ma, J. Genome sequencing of Streptomyces atratus SCSIO ZH16 and activation production of nocardamine via metabolic engineering. Front. Microbiol. 2018, 9, 1269. [Google Scholar] [CrossRef]
- Sun, C.; Yang, Z.; Zhang, C.; Liu, Z.; He, J.; Liu, Q.; Zhang, T.; Ju, J.; Ma, J. Genome Mining of Streptomyces atratus SCSIO ZH16: Discovery of Atratumycin and Identification of Its Biosynthetic Gene Cluster. Org. Lett. 2019, 21, 1453–1457. [Google Scholar] [CrossRef]
- Zhang, H.; White-Phillip, J.A.; Melançon, C.E.; Kwon, H.J.; Yu, W.L.; Liu, H.W. Elucidation of the Kijanimicin Gene Cluster: Insights into the Biosynthesis of Spirotetronate Antibiotics and Nitrosugars. J. Am. Chem. Soc. 2007, 129, 14670–14683. [Google Scholar] [CrossRef] [Green Version]
- Fang, J.; Zhang, Y.; Huang, L.; Jia, X.; Zhang, Q.; Zhang, X.; Tang, G.; Liu, W. Cloning and Characterization of the Tetrocarcin A Gene Cluster from Micromonospora chalcea NRRL 11289 Reveals a Highly Conserved Strategy for Tetronate Biosynthesis in Spirotetronate Antibiotics. J. Bacteriol. 2008, 190, 6014–6025. [Google Scholar] [CrossRef] [Green Version]
- Wei, R.B.; Xi, T.; Li, J.; Wang, P.; Li, F.C.; Lin, Y.C.; Qin, S. Lobophorin C and D, New Kijanimicin Derivatives from a Marine Sponge-Associated Actinomycetal Strain AZS17. Mar. Drugs 2011, 9, 359–368. [Google Scholar] [CrossRef]
- Niu, S.; Li, S.; Chen, Y.; Tian, X.; Zhang, H.; Zhang, G.; Zhang, W.; Yang, X.; Zhang, S.; Ju, J.; et al. Lobophorins E and F, new spirotetronate antibiotics from a South China Sea-derived Streptomyces sp. SCSIO 01127. J. Antibiolt. 2011, 64, 711. [Google Scholar] [CrossRef]
- Li, S.; Xiao, J.; Zhu, Y.; Zhang, G.; Yang, C.; Zhang, H.; Ma, L.; Zhang, C. Dissecting Glycosylation Steps in Lobophorin Biosynthesis Implies an Iterative Glycosyltransferase. Org. Lett. 2013, 15, 1374–1377. [Google Scholar] [CrossRef] [PubMed]
- Pan, H.Q.; Zhang, S.Y.; Wang, N.; Li, Z.L.; Hua, H.M.; Hu, J.C.; Wang, S.J. New Spirotetronate Antibiotics, Lobophorins H and I, from a South China Sea-Derived Streptomyces sp. 12A35. Mar. Drugs 2013, 11, 3891–3901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.; Wang, J.; Guo, H.; Hou, W.; Yang, N.; Ren, B.; Liu, M.; Dai, H.; Liu, X.; Song, F.; et al. Three antimycobacterial metabolites identified from a marine-derived Streptomyces sp. MS100061. Appl. Microbiol. Biotechnol. 2013, 97, 3885–3892. [Google Scholar] [CrossRef] [PubMed]
- Cruz, P.G.; Fribley, A.M.; Miller, J.R.; Larsen, M.J.; Schultz, P.J.; Jacob, R.T.; Tamayo-Castillo, G.; Kaufman, R.J.; Sherman, D.H. Novel Lobophorins Inhibit Oral Cancer Cell Growth and Induce Atf4- and Chop-Dependent Cell Death in Murine Fibroblasts. ACS Med. Chem. Lett. 2015, 6, 877–881. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Pan, H.; Hu, J. Isolation and identification of a new antibiotic, lobophorin J, from a deep sea-derived Streptomyces sp. 12A35. Chin. J. Antibiot. 2015, 40, 721–727. [Google Scholar]
- Yue, C.; Niu, J.; Liu, N.; Lü, Y.; Liu, M.; Li, Y. Cloning and identification of the lobophorin biosynthetic gene cluster from marine Streptomyces olivaceus strain FXJ7.023. Pak. J. Pharm. Sci. 2016, 29, 287–293. [Google Scholar] [PubMed]
- Braña, A.; Sarmiento-Vizcaíno, A.; Osset, M.; Pérez-Victoria, I.; Martín, J.; de Pedro, N.; de la Cruz, M.; Díaz, C.; Vicente, F.; Reyes, F.; et al. Lobophorin K, a new natural product with cytotoxic activity produced by Streptomyces sp. M-207 associated with the deep-sea coral Lophelia pertusa. Mar. Drugs. 2017, 15, 144. [Google Scholar] [CrossRef]
- Low, Z.J.; Pang, L.M.; Ding, Y.; Cheang, Q.W.; Hoang, K.L.M.; Tran, H.T.; Li, J.; Liu, X.-W.; Kanagasundaram, Y.; Yang, L.; et al. Identification of a biosynthetic gene cluster for the polyene macrolactam sceliphrolactam in a Streptomyces strain isolated from mangrove sediment. Sci. Rep. 2018, 8, 1594. [Google Scholar] [CrossRef]
- Zhang, C.; Sun, C.; Huang, H.; Gui, C.; Wang, L.; Li, Q.; Ju, J. Biosynthetic Baeyer–Villiger Chemistry Enables Access to Two Anthracene Scaffolds from a Single Gene Cluster in Deep-Sea-Derived Streptomyces olivaceus SCSIO T05. J. Nat. Prod. 2018, 81, 1570–1577. [Google Scholar] [CrossRef]
- Chin, C.-S.; Alexander, D.H.; Marks, P.; Klammer, A.A.; Drake, J.; Heiner, C.; Clum, A.; Copeland, A.; Huddleston, J.; Eichler, E.E.; et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 2013, 10, 563–569. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. AntiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef] [PubMed]
- Scherlach, K.; Hertweck, C. Triggering cryptic natural product biosynthesis in microorganisms. Org. Biomol. Chem. 2009, 7, 1753. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Zhang, C.; Qin, X.; Wei, X.; Liu, Q.; Li, Q.; Ju, J. Genome mining of Streptomyces olivaceus SCSIO T05: Discovery of olimycins A and B and assignment of absolute configurations. Tetrahedron 2018, 74, 199–203. [Google Scholar] [CrossRef]
- Zhang, C.; Yang, Z.; Qin, X.; Ma, J.; Sun, C.; Huang, H.; Li, Q.; Ju, J. Genome Mining for Mycemycin: Discovery and Elucidation of Related Methylation and Chlorination Biosynthetic Chemistries. Org. Lett. 2018, 20, 7633–7636. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Shen, B. Genes for Production of the Enediyne Antitumor Antibiotic C-1027 in Streptomyces globisporus Are Clustered with the cagA Gene That Encodes the C-1027 Apoprotein. Antimicrob. Agents Chemother. 2000, 44, 382–392. [Google Scholar] [CrossRef] [PubMed]
- Gust, B.; Challis, G.L.; Fowler, K.; Kieser, T.; Chater, K.F. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. USA 2003, 100, 1541–1546. [Google Scholar] [CrossRef] [PubMed]





| Feature | Value |
|---|---|
| Genome size (bp) | 8,458,055 |
| Average GC content (%) | 72.51 |
| Protein-coding genes | 7700 |
| Total size of Protein-coding genes (bp) | 7,543,173 |
| rRNAs number | 18 |
| tRNAs number | 65 |
| BGC | Position | Type (Product) | |
|---|---|---|---|
| From | To | ||
| Cluster 1 | 2725 | 89768 | Type I Polyketide synthase (T1 PKS) |
| Cluster 2 | 234616 | 284137 | Non-ribosomal peptide synthetase (NRPS) cluster |
| Cluster 3 | 504553 | 512728 | Bacteriocin or other unspecified ribosomally synthesized and post-translationally modified peptide product (RiPP) cluster (Bacteriocin) |
| Cluster 4 | 525945 | 544617 | Terpene |
| Cluster 5 | 793277 | 855894 | NRPS |
| Cluster 6 | 901333 | 979368 | T1 PKS |
| Cluster 7 | 980891 | 1005613 | Lanthipeptide cluster (Lanthipeptide) |
| Cluster 8 | 1135886 | 1240760 | Other types of PKS cluster (Otherks)-NRPS |
| Cluster 9 | 1275164 | 1347740 | NRPS-Terpene |
| Cluster 10 | 1651711 | 1694648 | NRPS-Nucleoside cluster (Nucleoside) |
| Cluster 11 | 1695020 | 1734380 | Otherks |
| Cluster 12 | 1751698 | 1796277 | NRPS |
| Cluster 13 | 1840051 | 1851963 | Siderophore cluster (Siderophore) |
| Cluster 14 | 1967451 | 1990613 | Lanthipeptide |
| Cluster 15 | 2037772 | 2059400 | Terpene |
| Cluster 16 | 2090680 | 2102023 | Bacteriocin |
| Cluster 17 | 2138860 | 2187226 | T1PKS-NRPS |
| Cluster 18 | 2230691 | 2317060 | NRPS-Type II PKS (T2 PKS)-Otherks |
| Cluster 19 | 2330735 | 2352337 | Lanthipeptide |
| Cluster 20 | 2443907 | 2456009 | Siderophore |
| Cluster 21 | 2905748 | 2978302 | T2 PKS |
| Cluster 22 | 3029068 | 3048760 | Terpene |
| Cluster 23 | 3049806 | 3075321 | Beta-lactone containing protease inhibitor (Betalactone) |
| Cluster 24 | 3182776 | 3235915 | NRPS |
| Cluster 25 | 3764472 | 3822515 | NRPS |
| Cluster 26 | 4131410 | 4159582 | Lanthipeptide |
| Cluster 27 | 4881296 | 4901736 | Phenazine cluster (Phenazine) |
| Cluster 28 | 5633979 | 5656500 | Lasso peptide cluster (Lassopeptide) |
| Cluster 29 | 5716930 | 5727556 | Melanin cluster (Melanin) |
| Cluster 30 | 6667385 | 6677783 | Ectoine cluster (Ectoine) |
| Cluster 31 | 7200930 | 7253804 | NRPS |
| Cluster 32 | 7328818 | 7368924 | Type III PKS (T3 PKS) |
| Cluster 33 | 7614814 | 7636052 | Aminoglycoside/aminocyclitol cluster (Amglyccycl) |
| Cluster 34 | 7882883 | 7906528 | Terpene |
| Cluster 35 | 7959695 | 7980831 | Indole cluster (Indole) |
| Cluster 36 | 8200560 | 8221618 | Terpene |
| Cluster 37 | 8239655 | 8455702 | T1pks-Nrps-T3 PKS-Oligosaccharide cluster (Oligosaccharide)-Other |
| ORF | Size a | Proposed Function | ID/SI b | Protein Homologue and Origin |
|---|---|---|---|---|
| orf(-2) | 374 | macrolide glycosyltransferase | 100/100 | Orf(-2) (AGI99472.1); Streptomyces sp. SCSIO 01127 |
| orf(-1) | 260 | FkbM family methyltransferase | 100/100 | Orf(-1) (AGI99473.1); Streptomyces sp. SCSIO 01127 |
| lbpR1 | 195 | TetR type regulatory protein | 100/100 | lobR1 (AGI99474.1); Streptomyces sp. SCSIO 01127 |
| lbpT1 | 497 | efflux permease | 100/100 | lobT1 (AGI99475.1); Streptomyces sp. SCSIO 01127 |
| lbpP1 | 392 | p450 monooxygenase | 100/100 | lobP1 (AGI99476.1); Streptomyces sp. SCSIO 01127 |
| lbpU1 | 326 | aldo/keto reductase | 100/100 | lobU1 (AGI99477.1); Streptomyces sp. SCSIO 01127 |
| lbpS1 | 271 | sugar-O-methyltransferase | 99/100 | lobS1 (AGI99478.1); Streptomyces sp. SCSIO 01127 |
| lbpS2 | 384 | sugar 4-aminotransferase | 100/100 | lobS2 (AGI99479.1); Streptomyces sp. SCSIO 01127 |
| lbpS3 | 201 | SAM-dependent methyltransferase | 97/98 | lobS3 (AGI99480.1); Streptomyces sp. SCSIO 01127 |
| lbpU2 | 197 | hypothetical protein | 100/100 | hypothetical protein (KMB22099.1); Klebsiella pneumoniae |
| lbpG1 | 391 | glycosyltransferase | 100/100 | lobG1 (AGI99481.1); Streptomyces sp. SCSIO 01127 |
| lbpA1 | 3936 | PKS (KS-AT-DH-ER-KR-ACP-KS-AT-DH-KR-ACP) | 100/100 | lobA1 (AGI99482.1); Streptomyces sp. SCSIO 01127 |
| lbpS4 | 483 | sugar 2,3-dehydratase | 100/100 | lobS4 (AGI99483.1); Streptomyces sp. SCSIO 01127 |
| lbpB | 253 | thioesterase | 100/100 | lobB (AGI99484.1); Streptomyces sp. SCSIO 01127 |
| lbpP2 | 313 | FAD-dependent oxidoreductase | 100/100 | part of lobP2 (AGI99485.1); Streptomyces sp. SCSIO 01127 |
| lbpG2 | 416 | glycosyltransferase | 99/100 | lobG2 (AGI99486.1); Streptomyces sp. SCSIO 01127 |
| lbpG3 | 476 | glycosyltransferase | 99/100 | lobG3 (AGI99487.1); Streptomyces sp. SCSIO 01127 |
| lbpC1 | 680 | hydrolase superfamily dihydrolipo-amide acyltransferase-like protein | 99/99 | lobC1 (AGI99489.1); Streptomyces sp. SCSIO 01127 |
| lbpC2 | 75 | ACP | 99/100 | lobC2 (AGI99490.1); Streptomyces sp. SCSIO 01127 |
| lbpC3 | 621 | FkbH-like protein | 99/100 | lobC3 (AGI99491.1); Streptomyces sp. SCSIO 01127 |
| lbpC4 | 342 | ketoacyl acyl carrier protein synthase III | 100/100 | lobC4 (AGI99492.1); Streptomyces sp. SCSIO 01127 |
| lbpP3 | 492 | FAD-dependent oxidoreductase | 100/100 | lobP3 (AGI99493.1); Streptomyces sp. SCSIO 01127 |
| lbpA2 | 1573 | PKS (KS-AT-KR-ACP) | 99/100 | lobA2 (AGI99494.1); Streptomyces sp. SCSIO 01127 |
| lbpA3 | 1798 | PKS (KS-AT-DH-KR-ACP) | 99/99 | lobA3 (AGI99495.1); Streptomyces sp. SCSIO 01127 |
| lbpA4 | 4376 | PKS (KR-ACP-KS-AT-DH-KR-ACP-KS-AT-DH-KR-ACP) | 100/100 | part of lobA4 (AGI99496.1); Streptomyces sp. SCSIO 01127 |
| lbpA5 | 2881 | PKS (KS-AT-DH-KR-ACP-KS-AT-DH) | 99/98 | part of lobA4 (AGI99496.1); Streptomyces sp. SCSIO 01127 |
| lbpA6 | 6362 | PKS (KS-AT-ACP-KS-AT-DH-KR-ACP-KS-AT-DH-KR-ACP-KS-AT-DH-KR-ACP) | 99/99 | lobA5 (AGI99497.1); Streptomyces sp. SCSIO 01127 |
| lbpU3 | 151 | unknown | 100/100 | lobU2 (AGI99498.1); Streptomyces sp. SCSIO 01127 |
| lbpS5 | 414 | sugar 3-C-methyl transferase | 100/100 | lobS5 (AGI99499.1); Streptomyces sp. SCSIO 01127 |
| lbpS6 | 373 | sugar 3-aminotransferase | 100/100 | lobS6 (AGI99500.1); Streptomyces sp. SCSIO 01127 |
| lbpS7 | 439 | acyl-CoA dehydrogenase | 100/100 | lobS7 (AGI99501.1); Streptomyces sp. SCSIO 01127 |
| lbpS8 | 341 | sugar 4,6-dehydratase | 100/100 | lobS8 (AGI99502.1); Streptomyces sp. SCSIO 01127 |
| lbpS9 | 298 | sugar nucleotidyltransferase | 99/100 | lobS9 (AGI99503.1); Streptomyces sp. SCSIO 01127 |
| lbpS10 | 332 | sugar 3-ketoreductase | 100/100 | lobS10 (AGI99504.1); Streptomyces sp. SCSIO 01127 |
| lbpS11 | 202 | sugar 5-epimerase | 99/100 | lobS11 (AGI99505.1); Streptomyces sp. SCSIO 01127 |
| lbpR2 | 274 | TetR type regulatory protein | 99/100 | lobR3 (AGI99506.1); Streptomyces sp. SCSIO 01127 |
| lbpT2 | 211 | forkhead-associated protein | 99/100 | lobT2 (AGI99507.1); Streptomyces sp. SCSIO 01127 |
| lbpR3 | 298 | putative regulatory protein | 99/100 | lobR4 (AGI99508.1); Streptomyces sp. SCSIO 01127 |
| lbpR4 | 309 | LysR family transcriptional regulator | 99/100 | lobR5 (AGI99509.1); Streptomyces sp. SCSIO 01127 |
| orf1 | 183 | acetyltransferase | 100/100 | Orf1 (AGI99510.1); Streptomyces sp. SCSIO 01127 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, C.; Ding, W.; Qin, X.; Ju, J. Genome Sequencing of Streptomyces olivaceus SCSIO T05 and Activated Production of Lobophorin CR4 via Metabolic Engineering and Genome Mining. Mar. Drugs 2019, 17, 593. https://doi.org/10.3390/md17100593
Zhang C, Ding W, Qin X, Ju J. Genome Sequencing of Streptomyces olivaceus SCSIO T05 and Activated Production of Lobophorin CR4 via Metabolic Engineering and Genome Mining. Marine Drugs. 2019; 17(10):593. https://doi.org/10.3390/md17100593
Chicago/Turabian StyleZhang, Chunyan, Wenjuan Ding, Xiangjing Qin, and Jianhua Ju. 2019. "Genome Sequencing of Streptomyces olivaceus SCSIO T05 and Activated Production of Lobophorin CR4 via Metabolic Engineering and Genome Mining" Marine Drugs 17, no. 10: 593. https://doi.org/10.3390/md17100593
APA StyleZhang, C., Ding, W., Qin, X., & Ju, J. (2019). Genome Sequencing of Streptomyces olivaceus SCSIO T05 and Activated Production of Lobophorin CR4 via Metabolic Engineering and Genome Mining. Marine Drugs, 17(10), 593. https://doi.org/10.3390/md17100593