In Silico Identification and Experimental Validation of (−)-Muqubilin A, a Marine Norterpene Peroxide, as PPARα/γ-RXRα Agonist and RARα Positive Allosteric Modulator
Abstract
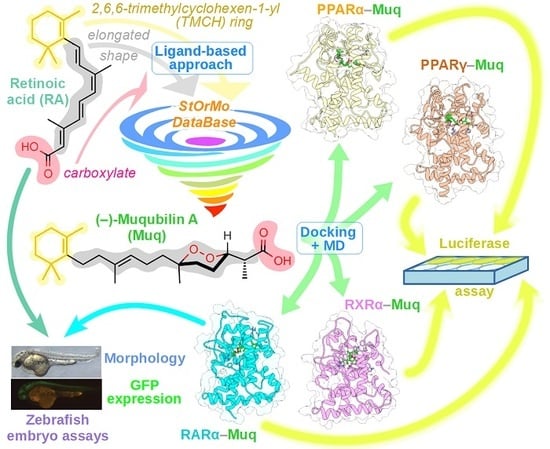
:1. Introduction
2. Results
2.1. Virtual Screening of Our In-House Molecular Database
2.2. Muq-RXRα Complex Model
2.3. Muq-PPARα/γ Complex Models
2.4. Muq-RARα Complex Model
2.5. Muq is an Agonist for RXRα and PPARα/γ
2.6. Muq Functions as an Allosteric Enhancer of RA on hRARα
3. Discussion and Conclusions
4. Materials and Methods
4.1. Computational Methods
4.2. Purification of (−)-Muqubilin A
4.3. Luciferase Assay
4.4. MTT Assay
4.5. Zebrafish Husbandry and Treatments
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Huang, P.; Chandra, V.; Rastinejad, F. Retinoic acid actions through mammalian nuclear receptors. Chem. Rev. 2014, 114, 233–254. [Google Scholar] [CrossRef] [PubMed]
- Sever, R.; Glass, C.K. Signaling by nuclear receptors. Cold Spring Harb. Perspect. Biol. 2013, 5, a016709. [Google Scholar] [CrossRef] [PubMed]
- Berger, J.; Moller, D.E. The Mechanisms of Action of PPARs. Annu. Rev. Med. 2002, 53, 409–435. [Google Scholar] [CrossRef]
- Altucci, L.; Leibowitz, M.D.; Ogilvie, K.M.; de Lera, A.R.; Gronemeyer, H. RAR and RXR modulation in cancer and metabolic disease. Nat. Rev. Drug Discov. 2007, 6, 793–810. [Google Scholar] [CrossRef] [PubMed]
- Glass, C.K.; Saijo, K. Nuclear receptor transrepression pathways that regulate inflammation in macrophages and T cells. Nat. Rev. Immunol. 2010, 10, 365–376. [Google Scholar] [CrossRef] [PubMed]
- Moutinho, M.; Landreth, G.E. Therapeutic potential of nuclear receptor agonists in Alzheimer’s disease. J. Lipid Res. 2017, 58, 1937–1949. [Google Scholar] [CrossRef]
- Uray, I.P.; Dmitrovsky, E.; Brown, P.H. Retinoids and rexinoids in cancer prevention: From laboratory to clinic. Semin. Oncol. 2016, 43, 49–64. [Google Scholar] [CrossRef]
- Zhang, H.; Xu, X.; Chen, L.; Chen, J.; Hu, L.; Jiang, H.; Shen, X. Molecular determinants of magnolol targeting both RXRα and PPARγ. PLoS ONE 2011, 6, e28253. [Google Scholar] [CrossRef]
- Bhatia, V.; Viswanathan, P. Insulin resistance and PPAR insulin sensitizers. Curr. Opin. Investig. Drugs 2006, 7, 891–897. [Google Scholar] [PubMed]
- Cramer, P.E.; Cirrito, J.R.; Wesson, D.W.; Lee, C.Y.D.; Karlo, J.C.; Zinn, A.E.; Casali, B.T.; Restivo, J.L.; Goebel, W.D.; James, M.J.; et al. ApoE-Directed Therapeutics Rapidly Clear-Amyloid and Reverse Deficits in AD Mouse Models. Science 2012, 335, 1503–1506. [Google Scholar] [CrossRef]
- Heneka, M.T.; Reyes-Irisarri, E.; Hull, M.; Kummer, M.P. Impact and therapeutic potential of PPARs in Alzheimers Disease. Curr. Neuropharmacol. 2011, 9, 643–650. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.J.; Kwon, K.J.; Park, J.Y.; Lee, S.H.; Moon, C.H.; Baik, E.J. Effects of peroxisome proliferator-activated receptor agonists on LPS-induced neuronal death in mixed cortical neurons: associated with iNOS and COX-2. Brain Res. 2002, 941, 1–10. [Google Scholar] [CrossRef]
- Corbett, G.T.; Gonzalez, F.J.; Pahan, K. Activation of peroxisome proliferator-activated receptor α stimulates ADAM10-mediated proteolysis of APP. Proc. Natl. Acad. Sci. 2015, 112, 8445–8450. [Google Scholar] [CrossRef] [PubMed]
- Husson, M.; Enderlin, V.; Delacourte, A.; Ghenimi, N.; Alfos, S.; Pallet, V.; Higueret, P. Retinoic acid normalizes nuclear receptor mediated hypo-expression of proteins involved in β-amyloid deposits in the cerebral cortex of vitamin A deprived rats. Neurobiol. Dis. 2006, 23, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Corcoran, J.P.T.; So, P.L.; Maden, M. Disruption of the retinoid signalling pathway causes a deposition of amyloid beta in the adult rat brain. Eur. J. Neurosci. 2004, 20, 896–902. [Google Scholar] [CrossRef] [PubMed]
- D’Aniello, E.; Fellous, T.; Iannotti, F.A.; Gentile, A.; Allarà, M.; Balestrieri, F.; Gray, R.; Amodeo, P.; Vitale, R.M.; Di Marzo, V. Identification and characterization of phytocannabinoids as novel dual PPARα/γ agonists by a computational and in vitro experimental approach. Biochim. Biophys. Acta Gen. Subj. 2019, 1863, 586–597. [Google Scholar] [CrossRef]
- Vitale, R.; D’Aniello, E.; Gorbi, S.; Martella, A.; Silvestri, C.; Giuliani, M.; Fellous, T.; Gentile, A.; Carbone, M.; Cutignano, A.; et al. Fishing for Targets of Alien Metabolites: A Novel Peroxisome Proliferator-Activated Receptor (PPAR) Agonist from a Marine Pest. Mar. Drugs 2018, 16, 431. [Google Scholar] [CrossRef]
- Vitale, R.M.; Rispoli, V.; Desiderio, D.; Sgammato, R.; Thellung, S.; Canale, C.; Vassalli, M.; Carbone, M.; Ciavatta, M.L.; Mollo, E.; et al. In Silico identification and experimental validation of novel anti-alzheimer’s multitargeted ligands from a marine source featuring a “2-aminoimidazole plus aromatic group” scaffold. ACS Chem. Neurosci. 2018, 9, 1290–1303. [Google Scholar] [CrossRef]
- Vitale, R.M.; Gatti, M.; Carbone, M.; Barbieri, F.; Felicità, V.; Gavagnin, M.; Florio, T.; Amodeo, P. Minimalist Hybrid Ligand/Receptor-Based Pharmacophore Model for CXCR4 Applied to a Small-Library of Marine Natural Products Led to the Identification of Phidianidine A as a New CXCR4 Ligand Exhibiting Antagonist Activity. ACS Chem. Biol. 2013, 8, 2762–2770. [Google Scholar] [CrossRef]
- Kashman, Y.; Rotem, M. Muqubilin, a new c24-isoprenoid from a marine sponge. Tetrahedron Lett. 1979, 20, 1707–1708. [Google Scholar] [CrossRef]
- Sperry, S.; Valeriote, F.A.; Corbett, T.H.; Crews, P. Isolation and cytotoxic evaluation of marine sponge-derived norterpene peroxides. J. Nat. Prod. 1998, 61, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Lefranc, F.; Nuzzo, G.; Hamdy, N.A.; Fakhr, I.; Moreno Y Banuls, L.; Van Goietsenoven, G.; Villani, G.; Mathieu, V.; van Soest, R.; Kiss, R.; et al. In vitro pharmacological and toxicological effects of norterpene peroxides isolated from the red sea sponge Diacarnus erythraeanus on normal and cancer cells. J. Nat. Prod. 2013, 76, 1541–1547. [Google Scholar] [CrossRef] [PubMed]
- Chao, C.-H.; Chou, K.-J.; Wang, G.-H.; Wu, Y.-C.; Wang, L.-H.; Chen, J.-P.; Sheu, J.-H.; Sung, P.-J. Norterpenoids and related peroxides from the formosan marine sponge negombata corticata. J. Nat. Prod. 2010, 73, 1538–1543. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, S.R.M.; Ebel, R.; Wray, V.; Müller, W.E.G.; Edrada-Ebel, R.; Proksch, P. Diacarperoxides, norterpene cyclic peroxides from the sponge diacarnus megaspinorhabdosa. J. Nat. Prod. 2008, 71, 1358–1364. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.S.; Desai-Yajnik, V.; Greene, M.E.; Raaka, B.M.; Samuels, H.H. The ligand-binding domains of the thyroid hormone/retinoid receptor gene subfamily function in vivo to mediate heterodimerization, gene silencing, and transactivation. Mol. Cell. Biol. 1995, 15, 1817–1825. [Google Scholar] [CrossRef] [PubMed]
- Cesario, R.M.; Stone, J.; Yen, W.-C.; Bissonnette, R.P.; Lamph, W.W. Differentiation and growth inhibition mediated via the RXR:PPARγ heterodimer in colon cancer. Cancer Lett. 2006, 240, 225–233. [Google Scholar] [CrossRef] [PubMed]
- Abba, M.C.; Hu, Y.; Levy, C.C.; Gaddis, S.; Kittrell, F.S.; Zhang, Y.; Hill, J.; Bissonnette, R.P.; Medina, D.; Brown, P.H.; et al. Transcriptomic signature of Bexarotene (Rexinoid LGD1069) on mammary gland from three transgenic mouse mammary cancer models. BMC Med. Genom. 2008, 1, 40. [Google Scholar] [CrossRef] [PubMed]
- D’Aniello, E.; Rydeen, A.B.; Anderson, J.L.; Mandal, A.; Waxman, J.S. Depletion of retinoic acid receptors initiates a novel positive feedback mechanism that promotes teratogenic increases in retinoic acid. PLoS Genet. 2013, 9, e1003689. [Google Scholar] [CrossRef]
- D’Aniello, E.; Ravisankar, P.; Waxman, J.S. Rdh10a provides a conserved critical step in the synthesis of retinoic acid during zebrafish embryogenesis. PLoS ONE 2015, 10, e0138588. [Google Scholar] [CrossRef]
- Mandal, A.; Rydeen, A.; Anderson, J.; Sorrell, M.R.J.; Zygmunt, T.; Torres-Vázquez, J.; Waxman, J.S. Transgenic retinoic acid sensor lines in zebrafish indicate regions of available embryonic retinoic acid. Dev. Dyn. 2013, 242, 989–1000. [Google Scholar] [CrossRef]
- Waxman, J.S.; Yelon, D. Zebrafish retinoic acid receptors function as context-dependent transcriptional activators. Dev. Biol. 2011, 352, 128–140. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Wang, R.-P.; Xu, B.; Yu, H.-B.; Ma, G.-Y.; Wang, G.-F.; Dai, S.-W.; Zhang, W.; Jiao, W.-H.; Song, S.-J.; et al. New antimalarial norterpene cyclic peroxides from Xisha Islands sponge Diacarnus megaspinorhabdosa. Bioorg. Med. Chem. Lett. 2016, 26, 2084–2087. [Google Scholar] [CrossRef] [PubMed]
- Hassinen, T.; Peräkylä, M. New energy terms for reduced protein models implemented in an Off-Lattice force field. J. Comput. Chem. 2001, 22, 1229–1242. [Google Scholar] [CrossRef]
- Clark, M.; Cramer, R.D.; Van Opdenbosch, N. Validation of the general purpose tripos 5.2 force field. J. Comput. Chem. 1989, 10, 982–1012. [Google Scholar] [CrossRef]
- Marenich, A.V.; Cramer, C.J.; Truhlar, D.G. Universal solvation model based on solute electron density and on a continuum model of the solvent defined by the bulk dielectric constant and atomic surface tensions. J. Phys. Chem. B 2009, 113, 6378–6396. [Google Scholar] [CrossRef] [PubMed]
- Fox, T.; Kollman, P.A. Application of the RESP Methodology in the Parametrization of Organic Solvents. J. Phys. Chem. B 1998, 102, 8070–8079. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. Software news and update AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Götz, A.W.; Williamson, M.J.; Xu, D.; Poole, D.; Le Grand, S.; Walker, R.C. Routine Microsecond Molecular Dynamics Simulations with AMBER on GPUs. 1. Generalized Born. J. Chem. Theory Comput. 2012, 8, 1542–1555. [Google Scholar] [CrossRef]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef]
- Ryckaert, J.-P.; Ciccotti, G.; Berendsen, H.J.C. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977, 23, 327–341. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Iannotti, F.A.; Panza, E.; Barrese, V.; Viggiano, D.; Soldovieri, M.V.; Taglialatela, M. Expression, localization, and pharmacological role of kv7 potassium channels in skeletal muscle proliferation, differentiation, and survival after myotoxic insults. J. Pharmacol. Exp. Ther. 2010, 332, 811–820. [Google Scholar] [CrossRef] [PubMed]




© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
D’Aniello, E.; Iannotti, F.A.; Falkenberg, L.G.; Martella, A.; Gentile, A.; De Maio, F.; Ciavatta, M.L.; Gavagnin, M.; Waxman, J.S.; Di Marzo, V.; et al. In Silico Identification and Experimental Validation of (−)-Muqubilin A, a Marine Norterpene Peroxide, as PPARα/γ-RXRα Agonist and RARα Positive Allosteric Modulator. Mar. Drugs 2019, 17, 110. https://doi.org/10.3390/md17020110
D’Aniello E, Iannotti FA, Falkenberg LG, Martella A, Gentile A, De Maio F, Ciavatta ML, Gavagnin M, Waxman JS, Di Marzo V, et al. In Silico Identification and Experimental Validation of (−)-Muqubilin A, a Marine Norterpene Peroxide, as PPARα/γ-RXRα Agonist and RARα Positive Allosteric Modulator. Marine Drugs. 2019; 17(2):110. https://doi.org/10.3390/md17020110
Chicago/Turabian StyleD’Aniello, Enrico, Fabio Arturo Iannotti, Lauren G. Falkenberg, Andrea Martella, Alessandra Gentile, Fabrizia De Maio, Maria Letizia Ciavatta, Margherita Gavagnin, Joshua S. Waxman, Vincenzo Di Marzo, and et al. 2019. "In Silico Identification and Experimental Validation of (−)-Muqubilin A, a Marine Norterpene Peroxide, as PPARα/γ-RXRα Agonist and RARα Positive Allosteric Modulator" Marine Drugs 17, no. 2: 110. https://doi.org/10.3390/md17020110
APA StyleD’Aniello, E., Iannotti, F. A., Falkenberg, L. G., Martella, A., Gentile, A., De Maio, F., Ciavatta, M. L., Gavagnin, M., Waxman, J. S., Di Marzo, V., Amodeo, P., & Vitale, R. M. (2019). In Silico Identification and Experimental Validation of (−)-Muqubilin A, a Marine Norterpene Peroxide, as PPARα/γ-RXRα Agonist and RARα Positive Allosteric Modulator. Marine Drugs, 17(2), 110. https://doi.org/10.3390/md17020110