The Purification, Characterization, and Biological Activity of New Polyketides from Mangrove-Derived Endophytic Fungus Epicoccum nigrum SCNU-F0002
Abstract
:1. Introduction
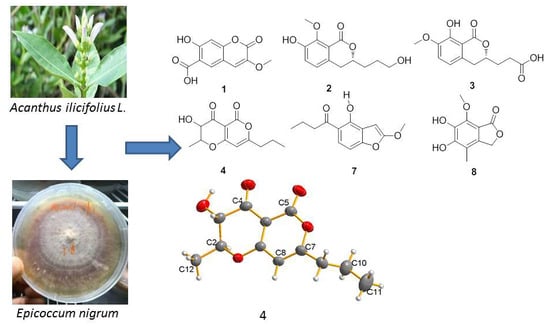
2. Results
3. Experimental Section
3.1. General Experimental Procedures
3.2. Fungal Materials
3.3. Extraction and Isolation
3.4. DPPH Radical Scavenging Activity Assay
3.5. Antimicrobial Activity Assay
3.6. Cytotoxicity Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gerwick, W.H.; Moore, B.S. Lessons from the past and charting the future of marine natural products drug discovery and chemical biology. Chem. Biol. 2012, 19, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Mostafa, E.R.; Rainer, E. Secondary metabolites of fungi from marine habitats. Nat. Prod. Rep. 2011, 28, 290–344. [Google Scholar]
- Blunt, J.W.; Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2018, 35, 8–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ancheeva, E.; Daletos, G.; Proksch, P. Lead compounds from mangrove-associated microorganisms. Mar. Drugs 2018, 16, 319. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Sun, B.; Gao, H.; Chen, X.; Liu, S.; Yao, X.; Liu, X.; Che, Y. Diketopiperazines from the cordyceps-colonizing fungus Epicoccum nigrum. J. Nat. Prod. 2009, 72, 2115–2119. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, S.; Che, Y.; Liu, X. Epicoccins A–D, Epipolythiodioxopiperazines from a cordyceps-colonizing isolate of Epicoccum nigrum. J. Nat. Prod. 2007, 70, 1522–1525. [Google Scholar] [CrossRef] [PubMed]
- Harwoko, H.; Daletos, G.; Stuhldreier, F.; Lee, J.; Weseelborg, S.; Feldbrugge, M.; Muller, W.E.G.; Kalscheuer, E.A.; Proksh, P. Dithiodiketopiperazine derivatives from endophytic fungi Trichoderma harzianum and Epicoccum nigrum. Nat. Prod. Res. 2019, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Gonda, S.; Kiss-Szikszai, A.; Szucs, Z.; Balla, B.; Vasas, G. Efficient biotransformation of non-steroid anti-inflammatory drugs by endophytic and epiphytic fungi from dried leaves of a medicinal plant, Plantago lanceolata L. Int. Biodeterior. Biodegrad. 2016, 108, 115–121. [Google Scholar] [CrossRef]
- Wang, J.M.; Ding, G.Z.; Fang, L.; Dai, J.G.; Yu, S.S.; Wang, Y.H.; Chen, X.G.; Ma, S.G.; Qu, J.; Xu, S.; et al. Thiodiketopiperazines produced by the endophytic fungus Epicoccum nigrum. J. Nat. Prod. 2010, 73, 1240–1249. [Google Scholar] [CrossRef]
- Cui, H.; Liu, Y.; Li, J.; Huang, X.; Yan, T.; Cao, W.; Liu, H.; Long, Y.; She, Z. Diaporindenes A–D: Four unusual 2, 3-dihydro-1H-indene analogues with anti-inflammatory activities from the mangrove endophytic fungus Diaporthe sp. SYSU-HQ3. J. Org. Chem. 2018, 83, 11804–11813. [Google Scholar] [CrossRef]
- Liu, Z.; Chen, S.; Qiu, P.; Tan, C.; Long, Y.; Lu, Y.; She, Z. (+)-and (−)-Ascomlactone A: A pair of novel dimeric polyketides from a mangrove endophytic fungus Ascomycota sp. SK2YWS-L. Org. Biomol. Chem. 2017, 15, 10276–10280. [Google Scholar] [CrossRef] [PubMed]
- Aldrich, T.J.; Rolshausen, P.E.; Roper, M.C.; Reader, J.M.; Steinhaus, M.J.; Rapicavoli, J.; Vosburg, D.A.; Maloney, K.N. Radicinin from Cochliobolus sp. inhibits Xylella fastidiosa, the causal agent of Pierce’s Disease of grapevine. Phytochemistry 2015, 116, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Kwak, J.H.; Lee, K.B.; Schmitz, F.J. Four new coumarin derivatives from Artemisia keiskeana. J. Nat. Prod. 2001, 64, 1081–1083. [Google Scholar] [CrossRef] [PubMed]
- Bai, M.; Zheng, C.J.; Huang, G.L.; Mei, R.Q.; Wang, B.; Luo, Y.P.; Zheng, C.; Niu, Z.G.; Chen, G.Y. Bioactive meroterpenoids and isocoumarins from the mangrove-derived fungus Penicillium sp. TGM112. J. Nat. Prod. 2019, 82, 1155–1164. [Google Scholar] [CrossRef] [PubMed]
- Choukchou-Braham, N.; Asakawa, Y.; Lepoittevin, J.P. Isolation, structure determination and synthesis of new dihydroisocoumarins from Ginkgo biloba L. Tetrahedron Lett. 1994, 35, 3949–3952. [Google Scholar] [CrossRef]
- Wangun, H.V.K.; Ishida, K.; Hertweck, C. Epicoccalone, a coumarin-type chymotrypsin inhibitor, and isobenzofuran congeners from an Epicoccum sp. associated with a tree fungus. Eur. J. Org. Chem. 2008, 2008, 3781–3784. [Google Scholar] [CrossRef]
- Chen, Q.F.; Zhou, M.; Yang, T.; Chen, X.Z.; Wang, C.; Zhang, G.L.; Li, G.Y. Secondary metabolites from fungus Alternaria sp. CIB 108. Chin. Chem. Lett. 2011, 22, 1226–1228. [Google Scholar]
- Trisuwan, K.; Rukachaisirikul, V.; Phongpaichit, S.; Preedanon, S.; Sakayaroj, J. Modiolide and pyrone derivatives from the sea fan-derived fungus Curvularia sp. PSU-F22. Arch. Pharm. Res. 2011, 34, 709–714. [Google Scholar] [CrossRef]
- Huang, X.Z.; Zhu, Y.; Guan, X.L.; Tian, K.; Guo, J.M.; Wang, H.B.; Fu, G.M. A novel antioxidant isobenzofuranone derivative from fungus Cephalosporium sp. AL031. Molecules 2012, 17, 4219–4224. [Google Scholar] [CrossRef]
- Lee, N.H.; Gloer, J.B.; Wicklow, D.T. Isolation of chromanone and isobenzofuran derivatives from a fungicolous isolate of Epicoccum purpurascens. Bull. Korean Chem. Soc. 2007, 28, 877–879. [Google Scholar] [CrossRef]
- Pang, X.; Lin, X.; Yang, J.; Zhou, X.; Yang, B.; Wang, J.; Liu, Y. Spiro-phthalides and isocoumarins isolated from the marine-sponge-derived fungus Setosphaeria sp. SCSIO41009. J. Nat. Prod. 2018, 81, 1860–1868. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Dong, M.; Chen, X.; Jiang, M.; Lv, X.; Zhou, J. Antimicrobial activity of an endophytic Xylaria sp. YX-28 and identification of its antimicrobial compound 7-amino-4-methylcoumarin. Appl. Microbiol. Biotechnol. 2008, 78, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Wang, R.; Zhao, W.; Yu, L.; Zhang, C.; Chang, S.; Li, Y.; Zhang, T.; Xing, J.; Gao, M.; et al. Isocoumarindole A, A chlorinated isocoumarin and indole alkaloid hybrid metabolite from an endolichenic fungus Aspergillus sp. Org. Lett. 2019, 21, 1530–1533. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Pei, S.; Ju, W.; Jia, M.; Tian, D.; Tang, Y.; Mao, G. Synthesis and in vitro and in vivo antitumour activity study of 11-hydroxyl esterified bergenin/cinnamic acid hybrids. Eur. J. Med. Chem. 2017, 133, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Giacomazza, D.; D’Andrea, D.; Provenzano, A.; Picone, P.; Provenzano, F.; Guarrasi, V.; Raimondo, M.; Biagio, P.L.S.; Passantino, R.; Mangione, M.R.; et al. The precious content of the olive mill wastewater: The protective effect of the antioxidant fraction in cell cultures. CyTA-J. Food 2018, 16, 658–666. [Google Scholar] [CrossRef]
- Chekuri, S.; Arunjyothi, B.; Anupalli, R.R. Radical scavenging activity (2,2-diphenyl-1-picrylhydrazyl) of acalypha indica linn. (Euphorbeace family). Int. J. Pharm. Sci. Res. 2018, 9, 313–317. [Google Scholar]
- Fernández-Sánchez, A.; Madrigal-Santillán, E.; Bautista, M.; Esquivel-Soto, J.; Morales-González, Á.; Esquivel-Chirino, C.; Durante-Montiel, I.; Sanchez-Rivera, G.; Valadez-Vega, C.; Morales-González, J.A. Inflammation, oxidative stress, and obesity. Int. J. Mol. Sci. 2011, 12, 3117–3132. [Google Scholar] [CrossRef] [PubMed]
- Young, I.S.; Woodside, J.V. Antioxidants in health and disease. J. Clin. Pathol. 2001, 54, 176–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nowak, D.; Gośliński, M.; Przygoński, K.; Wojtowicz, E. The antioxidant properties of exotic fruit juices from acai, maqui berry and noni berries. Eur. Food Res. Technol. 2018, 244, 1897–1905. [Google Scholar] [CrossRef]
- Chen, S.; Liu, Y.; Liu, Z. Isocoumarins and benzofurans from the mangrove endophytic fungus Talaromyces amestolkiae possess α-glucosidase inhibitory and antibacterial activities. RSC Adv. 2016, 6, 26412–26420. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, Q.; Xia, G.; Huang, H.; Li, H.; Ma, L.; Lu, Y.; He, L.; Xia, X.; She, Z. Polyketides with α-glucosidase inhibitory activity from a mangrove endophytic fungus, Penicillium sp. HN29-3B1. J. Nat. Prod. 2015, 78, 1816–1822. [Google Scholar] [CrossRef] [PubMed]
- Hooft, R.W.; Straver, L.H.; Spek, A.L. Determination of absolute structure using Bayesian statistics on Bijvoet differences. J. Appl. Cryst. 2008, 41, 96–103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spek, A.L. Structure validation in chemical crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 2009, 65, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Robeson, D.J.; Gray, G.R.; Strobel, G.A. Production of the phytotoxins radicinin and radicinol by Alternaria chrysanthemi. Phytochemistry 1982, 21, 2359–2362. [Google Scholar] [CrossRef]
- Sugiyama, Y.; Ito, Y.; Suzuki, M.; Hirota, A. Indole derivatives from a marine sponge-derived yeast as DPPH radical scavengers. J. Nat. Prod. 2009, 72, 2069–2071. [Google Scholar] [CrossRef] [PubMed]
- Sharma, O.P.; Bhat, T.K. DPPH antioxidant assay revisited. Food Chem. 2008, 113, 1202–1205. [Google Scholar] [CrossRef]
- Carpinella, M.C.; Ferrayoli, C.G.; Palacios, S.M. Antifungal synergistic effect of scopoletin, a hydroxycoumarin isolated from Melia azedarach L. fruits. J. Agric. Food Chem. 2005, 53, 2922–2927. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Xie, Z.; Wu, M.; Li, X.; Li, W.; Ding, W.; She, Z.; Li, C. New antimicrobial cyclopentenones from Nigrospora sphaerica ZMT05, a fungus derived from Oxya chinensis thunber. J. Agric. Food Chem. 2018, 66, 5368–5372. [Google Scholar] [CrossRef]
- Chen, S.H.; Chen, D.N.; Cai, R.L.; Cui, H.; Long, Y.H.; Lu, Y.J.; Li, C.Y.; She, Z.G. Cytotoxic and antibacterial preussomerins from the mangrove endophytic fungus Lasiodiplodia theobromae ZJ-HQ1. J. Nat. Prod. 2016, 79, 2397–2402. [Google Scholar] [CrossRef]




| Position | 1 a | 2 b | 3 b | |||
|---|---|---|---|---|---|---|
| δC | δH(J/Hz) | δC | δH(J/Hz) | δC | δH(J/Hz) | |
| 1 | 165.2, C | 164.9, C | ||||
| 2 | 156.3, C | |||||
| 3 | 142.0, C | 80.3, CH | 4.48, m | 79.5, CH | 4.50, m | |
| 4 | 113.7, CH | 7.40, s | 34.3, CH2 | 2.88, m | 34.4, CH2 | 2.88, m |
| 4a | 112.4, C | 133.2, C | 133.0, C | |||
| 5 | 129.4, CH | 8.10, s | 124.3, CH | 6.95, d (8.2) | 124.3, CH | 6.94, d (8.2) |
| 6 | 111.0, C | 123.0, CH | 7.10, d (8.2) | 123.0, CH | 7.10, d (8.2) | |
| 7 | 153.8, C | 151.5, C | 150.5, C | |||
| 8 | 103.3, CH | 6.90, s | 150.5, C | 151.5, C | ||
| 8a | 161.2, C | 119.4, C | 119.3, C | |||
| 9 | 171.1, C | 32.2, CH2 | 1.83, m | 31.0, CH2 | 2.05, m | |
| 10 | 56.1, OCH3 | 3.79, s | 29.1, CH2 | 1.69, m | 30.6, CH2 | 2.54, m |
| 11 | 62.5, CH2 | 3.63, t (6.0) | 177.1, C | |||
| 12 | 61.9, OCH3 | 3.87, s | 61.9, OCH3 | 3.86, s | ||
| Position | 4 a | 7 b | 8 c | |||
|---|---|---|---|---|---|---|
| δC | δH(J/Hz) | δC | δH(J/Hz) | δC | δH(J/Hz) | |
| 1 | 173.4, C | |||||
| 2 | 80.2, CH | 4.36, dq (6, 12.6) | 162.9, C | |||
| 3 | 72.1, CH | 3.99, d (12.6) | 91.5, CH | 6.21, s | 70.1, CH2 | 5.17, s |
| 4 | 188.9, C | 166.2, C | 111.4, C | |||
| 4a | 97.6, C | 105.5, C | 143.8, C | |||
| 5 | 157.8, C | 104.1, C | 156.7, C | |||
| 6 | 124.3, CH | 7.09, d (13.2) | 149.2, C | |||
| 7 | 173.5, C | 119.8, CH | 7.15, d (13.2) | 136.6, C | ||
| 7a | 163.5, C | 103.9, C | ||||
| 8 | 99.1, CH | 5.90, s | 206, C | 10.7, CH3 | 2.06, s | |
| 8a | 176.5, C | |||||
| 9 | 36.5, CH2 | 2.48, t (7.2) | 46.7, CH2 | 2.98, t (7.2) | 61.2, OCH3 | 3.80, s |
| 10 | 19.9, CH2 | 1.71, qt (7.2, 7.8) | 18.8, CH2 | 1.67, qt (7.2, 7.8) | ||
| 11 | 13.5, CH3 | 0.98, t (7.8) | 14.2, CH3 | 0.96, t (7.8) | ||
| 12 | 18.2, CH3 | 1.64, d (6) | 56.2, OCH3 | 3.91, s | ||
| 4-OH | 15.11, s | |||||
| Strains | MIC (μg/mL) | |||||
|---|---|---|---|---|---|---|
| Compounds a | S. aureus (G+) | B. subtilis (G+) | E. coli (G−) | P. aeruginosa (G−) | S. enteritidis (G−) | |
| 6 | >100 | 50 | >100 | >100 | >100 | |
| 7 | >100 | 25 | 50 | >100 | >100 | |
| Ciprofloxacin b | 0.25 | 0.25 | 0.5 | 0.5 | 0.25 | |
| Compound | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | Vitamin C a |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IC50(μg/mL) | - | - | - | - | - | - | - | - | 62.9 | 13.6 | 12.1 | 18.1 | 11.7 | 18.2 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, Z.; Wen, S.; Ding, M.; Guo, H.; Huang, C.; Zhu, X.; Huang, J.; She, Z.; Long, Y. The Purification, Characterization, and Biological Activity of New Polyketides from Mangrove-Derived Endophytic Fungus Epicoccum nigrum SCNU-F0002. Mar. Drugs 2019, 17, 414. https://doi.org/10.3390/md17070414
Yan Z, Wen S, Ding M, Guo H, Huang C, Zhu X, Huang J, She Z, Long Y. The Purification, Characterization, and Biological Activity of New Polyketides from Mangrove-Derived Endophytic Fungus Epicoccum nigrum SCNU-F0002. Marine Drugs. 2019; 17(7):414. https://doi.org/10.3390/md17070414
Chicago/Turabian StyleYan, Zhangyuan, Shitong Wen, Meng Ding, Huixian Guo, Cuiying Huang, Xintong Zhu, Junyi Huang, Zhigang She, and Yuhua Long. 2019. "The Purification, Characterization, and Biological Activity of New Polyketides from Mangrove-Derived Endophytic Fungus Epicoccum nigrum SCNU-F0002" Marine Drugs 17, no. 7: 414. https://doi.org/10.3390/md17070414
APA StyleYan, Z., Wen, S., Ding, M., Guo, H., Huang, C., Zhu, X., Huang, J., She, Z., & Long, Y. (2019). The Purification, Characterization, and Biological Activity of New Polyketides from Mangrove-Derived Endophytic Fungus Epicoccum nigrum SCNU-F0002. Marine Drugs, 17(7), 414. https://doi.org/10.3390/md17070414