New Benzofuranoids and Phenylpropanoids from the Mangrove Endophytic Fungus, Aspergillus sp. ZJ-68
Abstract
:1. Introduction
2. Results
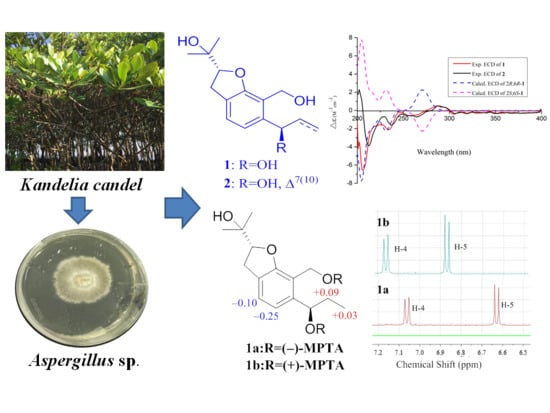
2.1. Structure Elucidation
2.2. Biological Activity
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Fungal Material
3.3. Fermentation
3.4. Extraction and Isolation
3.5. Inhibitory Activity Against α-Glucosidase
3.6. Antibacterial Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Wu, J.; Xiao, Q.; Xu, J.; Li, M.Y.; Pan, J.Y.; Yang, M.H. Natural products from true mangrove flora: Source, chemistry and bioactivities. Nat. Prod. Rep. 2008, 25, 955–981. [Google Scholar] [CrossRef] [PubMed]
- Thatoi, H.; Behera, B.C.; Mishra, R.R.; Dutta, S.K. Biodiversity and biotechnological potential of microorganisms from mangrove ecosystems: Areview. Ann. Microbiol. 2013, 63, 1–19. [Google Scholar] [CrossRef]
- Kusari, S.; Spiteller, M. Are we ready for industrial production of bioactive plant secondary metabolites utilizing endophytes. Nat. Prod. Rep. 2011, 28, 1203–1207. [Google Scholar] [CrossRef] [PubMed]
- Aly, A.H.; Debbab, A.; Proksch, P. Fungal endophytes: unique plant inhabitants with great promises. Appl. Microbiol. Biotechnol. 2011, 90, 1829–1845. [Google Scholar] [CrossRef] [PubMed]
- Ebrahim, W.; El-Neketi, M.; Lewald, L.I.; Orfali, R.S.; Lin, W.; Rehberg, N.; Kalscheuer, R.; Daletos, G.; Proksch, P. Metabolites from the fungal endophyte Aspergillus austroafricanus in axenic culture and in fungal–bacterial mixed cultures. J. Nat. Prod. 2016, 79, 914–922. [Google Scholar] [CrossRef] [PubMed]
- Frisvad, J.C.; Larsen, T.O. Extrolites of Aspergillus fumigatus and other pathogenic species in Aspergillus section Fumigati. Front. Microbiol. 2016, 6, 1485. [Google Scholar] [CrossRef] [PubMed]
- Lubertozzi, D.; Keasling, J.D. Developing Aspergillus as a host for heterologous expression. Biotechnol. Adv. 2009, 27, 53–75. [Google Scholar] [CrossRef] [PubMed]
- Novak, N.; Gerdin, S.; Berovic, M. Increased lovastatin formation by Aspergillus terreus using repeated fed-batch process. Biotechnol. Lett. 1997, 19, 947–948. [Google Scholar] [CrossRef]
- Chen, S.; Chen, D.; Cai, R.; Cui, H.; Long, Y.; Lu, Y.; Li, C.; She, Z. Cytotoxic and antibacterial preussomerins from the mangrove endophytic fungus Lasiodiplodia theobromae ZJ-HQ1. J. Nat. Prod. 2016, 79, 2397–2402. [Google Scholar] [CrossRef]
- Cui, H.; Liu, Y.; Nie, Y.; Liu, Z.; Chen, S.; Zhang, Z.; Lu, Y.; He, L.; Huang, X.; She, Z. Polyketides from the mangrove-derived endophytic fungus Nectria sp. HN001 and their α-glucosidase inhibitory activity. Mar. Drugs 2016, 14, 86. [Google Scholar] [CrossRef]
- Cui, H.; Lin, Y.; Luo, M.; Lu, Y.; Huang, X.; She, Z. Diaporisoindoles A–C: three isoprenylisoindole alkaloid derivatives from the mangrove endophytic fungus Diaporthe sp. SYSU-HQ3. Org. Lett. 2017, 19, 5621–5624. [Google Scholar] [CrossRef] [PubMed]
- Cai, R.; Wu, Y.; Chen, S.; Cui, H.; Liu, Z.; Li, C.; She, Z. Peniisocoumarins A–J: isocoumarins from Penicillium commune QQF-3, an endophytic fungus of the mangrove plant Kandelia candel. J. Nat. Prod. 2018, 81, 1376–1383. [Google Scholar] [CrossRef] [PubMed]
- Trisuwan, K.; Rukachaisirikul, V.; Sukpondma, Y.; Phongpaichit, S.; Preedanon, S.; Sakayaroj, J. Furo [3,2-h] isochroman, furo [3,2-h] isoquinoline, isochroman, phenol, pyranone, and pyrone derivatives from the sea fan-derived fungus Penicillium sp. PSU-F40. Tetrahedron 2010, 66, 4484–4489. [Google Scholar] [CrossRef]
- Kuramochi, K.; Tsubaki, K. Synthesis and structural characterization of natural benzofuranoids. J. Nat. Prod. 2015, 78, 1056–1066. [Google Scholar] [CrossRef]
- Xiao, Z.; Chen, S.; Cai, R.; Lin, S.; Hong, K.; She, Z. New furoisocoumarins and isocoumarins from the mangrove endophytic fungus Aspergillus sp. 085242. Beilstein J. Org. Chem. 2016, 12, 2077–2085. [Google Scholar] [CrossRef] [PubMed]
- Weber, D.; Gorzalczany, S.; Martino, V.; Acevedo, C.; Sterner, O.; Anke, T. Metabolites from endophytes of the medicinal plant Erythrina crista-galli. Z. Naturforsch. C 2005, 60, 467–477. [Google Scholar] [CrossRef]
- Bunbamrung, N.; Intaraudom, C.; Boonyuen, N.; Rachtawee, P.; Laksanacharoen, P.; Pittayakhajonwut, P. Penicisochromans from the endophytic fungus Penicillium sp. BCC18034. Phytochem. Lett. 2014, 10, 13–18. [Google Scholar] [CrossRef]
- Ayer, W.A.; Pena-Rodriguez, L.M. Metabolites produced by Alternaria brassicae, the black spot pathogen of canola. part 2, sesquiterpenoid metabolites. J. Nat. Prod. 1987, 50, 408–417. [Google Scholar] [CrossRef]
- Ogawa, A.; Murakami, C.; Kamisuki, S.; Kuriyama, I.; Yoshida, H.; Sugawara, F.; Mizushina, Y. Pseudodeflectusin, a novel isochroman derivative from Aspergillus pseudodeflectus a parasite of the sea weed, Sargassum fusiform, as a selective human cancer cytotoxin. Bioorg. Med. Chem. Lett. 2004, 14, 3539–3543. [Google Scholar] [CrossRef]
- Lu, Z.; Wang, Y.; Miao, C.; Liu, P.; Hong, K.; Zhu, W. Sesquiterpenoids and benzofuranoids from the marine-derived fungus Aspergillus ustus 094102. J. Nat. Prod. 2009, 72, 1761–1767. [Google Scholar] [CrossRef]
- Ohtani, I.; Kusumi, T.; Kashman, Y.; Kakisawa, H. High-field FT NMR application of Mosher’s method. The absolute configurations of marine terpenoids. J. Am. Chem. Soc. 1991, 113, 4092–4096. [Google Scholar] [CrossRef]
- Kjer, J.; Debbab, A.; Aly, A.H.; Proksch, P. Methods for isolation of marine-derived endophytic fungi and their bioactive secondary products. Nat. Protoc. 2010, 5, 479–490. [Google Scholar] [CrossRef]
- Cai, R.; Chen, S.; Liu, Z.; Tan, C.; Huang, X.; She, Z. A new α-pyrone from the mangrove endophytic fungus Phomopsis sp. HNY29-2B. Nat. Prod. Res. 2017, 31, 124–130. [Google Scholar] [CrossRef]





| No. | 1 | 2 | 3 | |||
|---|---|---|---|---|---|---|
| δC, Type | δH, mult (J/Hz) | δC, Type | δH, mult (J/Hz) | δC, Type | δH, mult (J/Hz) | |
| 2 | 89.6, CH | 4.57, t (9.0) | 89.6, CH | 4.60, t (8.9) | 89.7, CH | 4.37, s |
| 3 | 30.7, CH2 | 3.19, dd (8.5, 15.7) 3.08, dd (9.5, 15.6) | 30.7, CH2 | 3.21, dd (8.4, 15.8) 3.11, dd (9.5, 15.8) | 199.9, C | |
| 3a | 126.6, C | 127.5, C | 119.7, C | |||
| 4 | 124.7, CH | 7.04, d (7.6) | 124.6, CH | 7.06, d (7.6) | 124.2, CH | 7.47, d (7.9) |
| 5 | 118.5, CH | 6.87, d (7.6) | 120.0, CH | 6.84, d (7.6) | 117.6, CH | 6.63, d (7.9) |
| 5a | 142.5, C | 141.0, C | 142.6, C | |||
| 6 | 72.6, CH | 4.74, m | 72.6, CH | 5.41, m | 101.7, CH | 5.69, s |
| 7 | 30.2, CH2 | 1.87, m 1.78, m | 139.3, CH | 6.13, ddd (4.6, 10.5, 17.5) | 160.7, C | |
| 9 | 55.9, CH2 | 4.74, d (12.0) 4.63, d (12.0) | 56.2, CH2 | 4.81, d (12.1) 4.69, d (12.1) | 62.7, CH2 | 5.28, d (13.2) 5.22, d (13.2) |
| 9a | 119.8, C | 120.2, C | 108.9, C | |||
| 9b | 158.6, C | 158.9, C | 167.9, C | |||
| 10 | 10.9, CH3 | 0.94, t (7.4) | 115.3, CH2 | 5.39, d (9.1) 5.27, d (10.5) | 20.1, CH3 | 1.98, s |
| 11 | 71.7, C | 71.8, C | 72.6, C | |||
| 12 | 24.3, CH3 | 1.14, s | 24.2, CH3 | 1.17, s | 24.1, CH3 | 1.20, s |
| 13 | 26.7, CH3 | 1.36, s | 26.6, CH3 | 1.36, s | 26.2, CH3 | 1.36, s |
| No. | 4 | 5 | ||
|---|---|---|---|---|
| δC, Type | δH, mult (J/Hz) | δC, Type | δH, mult (J/Hz) | |
| 2 | 145.5, C | 145.5, C | ||
| 3 | 183.6, C | 183.6, C | ||
| 3a | 121.3, C | 121.3, C | ||
| 4 | 122.1, CH | 7.53, d (7.9) | 122.1, CH | 7.53, d (7.9) |
| 5 | 123.2, CH | 6.84, d (7.9) | 123.2, CH | 6.84, d (7.9) |
| 5a | 140.8, C | 140.9, C | ||
| 6 | 39.5, CH2 | 3.00, d (16.2) 2.92, d (16.2) | 39.4, CH2 | 3.00, d (16.5) 2.92, d (16.5) |
| 7 | 97.6, C | 97.6, C | ||
| 9 | 57.9, CH2 | 4.93, d (15.6) 4.72, d (15.6) | 57.9, CH2 | 4.93, d (15.6) 4.72, d (15.6) |
| 9a | 118.4, C | 118.4, C | ||
| 9b | 160.5, C | 160.5, C | ||
| 10 | 23.1, CH3 | 1.53, s | 23.1, CH3 | 1.53, s |
| 11 | 131.8, C | 131.8, C | ||
| 12 | 17.6, CH3 | 2.36, s | 17.6, CH3 | 2.36, s |
| 13 | 20.3, CH3 | 2.09, s | 20.3, CH3 | 2.09, s |
| 7-OCH3 | 49.2, CH3 | 3.34, s | 49.2, CH3 | 3.34, s |
| No | 6a | 7b | ||
|---|---|---|---|---|
| δC, Type | δH, mult (J/Hz) | δC, Type | δH, mult (J/Hz) | |
| 1 | 144.1, C | 111.0, CH | 6.93, d (8.3) | |
| 2 | 114.7, CH | 6.66, d (8.1) | 129.7, CH | 7.25, t (7.9) |
| 3 | 121.1, CH | 6.51, d (8.1) | 122.8, CH | 6.74, d (7.6) |
| 4 | 132.6, C | 139.0, C | ||
| 5 | 125.2, C | 127.7, C | ||
| 6 | 145.3, C | 159.6, C | ||
| 7 | 35.2, CH2 | 2.50, m | 130.2, CH | 6.79, d (11.5) |
| 8 | 25.6, CH2 | 1.51, m | 133.4, CH | 5.94, dt (6.7, 11.5) |
| 9 | 14.2, CH3 | 0.91, t (7.3) | 59.5, CH2 | 4.10, dd (1.3, 6.8) |
| 10 | 59.2, CH2 | 4.83, s | 56.6, CH2 | 4.64, s |
| 6-OCH3 | 56.1, CH3 | 3.85, s | ||
| Compounds a | % Inhibition (100 μM) | IC50 (μM) |
|---|---|---|
| 6 | 98 | 12.4 ± 1.0 |
| Acarbose b | 19 | 453.3 ± 1.0 |
| Compounds a | MIC (μg/mL) | |||
|---|---|---|---|---|
| S. aureus | E. coli | B. subtilis | P. aeruginosa | |
| 8 | 4.15 ± 1.12 | 8.3 ± 1.0 | 8.3 ± 1.1 | >100 |
| 11 | 12.5 ± 1.1 | 12.5 ± 1.2 | 12.5 ± 1.0 | >100 |
| ciprofloxacinb | 1.25 ± 1.10 | 1.25 ± 1.12 | 2.5 ± 1.1 | 2.5 ± 1.2 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cai, R.; Jiang, H.; Zang, Z.; Li, C.; She, Z. New Benzofuranoids and Phenylpropanoids from the Mangrove Endophytic Fungus, Aspergillus sp. ZJ-68. Mar. Drugs 2019, 17, 478. https://doi.org/10.3390/md17080478
Cai R, Jiang H, Zang Z, Li C, She Z. New Benzofuranoids and Phenylpropanoids from the Mangrove Endophytic Fungus, Aspergillus sp. ZJ-68. Marine Drugs. 2019; 17(8):478. https://doi.org/10.3390/md17080478
Chicago/Turabian StyleCai, Runlin, Hongming Jiang, Zhenming Zang, Chunyuan Li, and Zhigang She. 2019. "New Benzofuranoids and Phenylpropanoids from the Mangrove Endophytic Fungus, Aspergillus sp. ZJ-68" Marine Drugs 17, no. 8: 478. https://doi.org/10.3390/md17080478
APA StyleCai, R., Jiang, H., Zang, Z., Li, C., & She, Z. (2019). New Benzofuranoids and Phenylpropanoids from the Mangrove Endophytic Fungus, Aspergillus sp. ZJ-68. Marine Drugs, 17(8), 478. https://doi.org/10.3390/md17080478