Characterization of Nme5-Like Gene/Protein from the Red Alga Chondrus Crispus
Abstract
:1. Introduction
2. Results
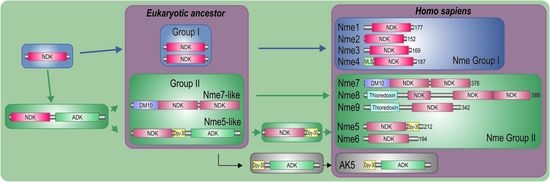
2.1. Nme Family in Eukaryotes
2.2. The Ancestral Nme5-LikeCc Protein Forms a Multimer
2.3. Recombinant Nme5-LikeCc Possesses NDPK Activity and Binds Nonspecifically to Various Types of DNA
2.4. The Ancestral Nme5-LikeCc Protein Localizes in Nucleus and Cytoplasm
2.5. The Ancestral Nme5-LikeCc Protein Diminished Colony Growth in HEK293T Cell Line and Inhibited Cell Proliferation
3. Discussion
4. Materials and Methods
4.1. Distribution of the Nme Protein Family Across Eukaryotic Supergroups and Phylogenetic Analyses
4.2. Isolation of Genomic DNA from Chondrus Crispus and Nme5-Like Gene Cloning
4.3. Expression and Purification of the Recombinant Nme5-LikeCc Protein and Western Analyses
4.4. Oligomerization of Protein and Gel Filtration Chromatography
4.5. NDP Kinase Assay
4.6. DNA-Binding Assay
4.7. Transient Transfections and Laser Scanning Confocal Microscopy
4.8. Soft Agar Colonization Assay
4.9. Cell Proliferation Assay
4.10. Detection of Apoptosis
4.11. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Steeg, P.S.; Bevilacqua, G.; Kopper, L.; Thorgeirsson, U.P.; Talmadge, J.E.; Liotta, L.A.; Sobel, M.E. Evidence for a novel gene associated with low tumor metastatic potential. J. Natl. Cancer Inst. 1988, 80, 200–204. [Google Scholar] [CrossRef] [PubMed]
- Desvignes, T.; Pontarotti, P.; Fauvel, C.; Bobe, J. Nme protein family evolutionary history, a vertebrate perspective. BMC Evol. Biol. 2009, 9, 256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoon, J.H.; Singh, P.; Lee, D.H.; Qiu, J.; Cai, S.; O’Connor, T.R.; Chen, Y.; Shen, B.; Pfeifer, G.P. Characterization of the 3′ → 5′ exonuclease activity found in human nucleoside diphosphate kinase 1 (NDK1) and several of its homologues. Biochemistry 2005, 44, 15774–15786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsuiki, H.; Nitta, M.; Furuya, A.; Hanai, N.; Fujiwara, T.; Inagaki, M.; Kochi, M.; Ushio, Y.; Saya, H.; Nakamura, H. A Novel Human Nucleoside Diphosphate (NDP) Kinase, Nm23-H6, Localizes in Mitochondria and Affects Cytokinesis. J. Cell. Biochem. 1999, 76, 254–269. [Google Scholar] [CrossRef]
- Herak Bosnar, M.; Radić, M.; Ćetković, H. A young researcher’s guide to NME/Nm23/NDP Kinase. Period. Biol. 2018, 120, 3–9. [Google Scholar] [CrossRef]
- Postel, E.H. Multiple biochemical activities of Nm23/NDP kinase in gene regulation. J Bioenerg. Biomembr. 2003, 35, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Hartsough, M.T.; Morrison, D.K.; Salerno, M.; Palmieri, D.; Ouatas, T.; Mair, M.; Patrick, J.; Steeg, P.S. Nm23-H1 metastasis suppressor phosphorylation of kinase suppressor of Ras via a histidine protein kinase pathway. J. Biol. Chem. 2002, 277, 32389–32399. [Google Scholar] [CrossRef] [Green Version]
- Fan, Z.; Beresford, P.J.; Oh, D.Y.; Zhang, D.; Lieberman, J. Tumor suppressor Nm23-H1 is a granzyme A-activated DNase during CTL-mediated apoptosis, and the nucleosome assembly protein SET is its inhibitor. Cell 2003, 112, 659–672. [Google Scholar] [CrossRef] [Green Version]
- Lee, M.Y.; Jeong, W.J.; Oh, J.W.; Choi, K.Y. Nm23H2 inhibits EGF- and Ras-induced proliferation of NIH3T3 cells by blocking the ERK pathway. Cancer Lett. 2009, 275, 221–226. [Google Scholar] [CrossRef]
- Gervasi, F.; D’Agnano, I.; Vossio, S.; Zupi, G.; Sacchi, A.; Lombardi, D. Nm23 influences proliferation and differentiation of PC12 cells in response to nerve growth factor. Cell Growth Differ. 1996, 7, 1689–1695. [Google Scholar]
- Masoudi, N.; Fancsalszky, L.; Pourkarimi, E.; Vellai, T.; Alexa, A.; Reményi, A.; Gartner, A.; Mehta, A.; Takács-Vellai, K. The NM23-H1/H2 homolog NDK-1 is required for full activation of Ras signaling in C. elegans. Development 2013, 140, 3486–3495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takacs-Vellai, K.; Vellai, T.; Farkas, Z.; Mehta, A. Nucleoside diphosphate kinases (NDPKs) in animal development. Cell. Mol. Life Sci. 2015, 72, 1447–1462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lombardi, D.; Palescandolo, E.; Giordano, A.; Paggi, M.G. Interplay between the antimetastatic Nm23 and the retinoblastoma-related Rb2/p130 genes in promoting neuronal differentiation of PC12 cells. Cell Death Differ. 2001, 8, 470–476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lombardi, D.; Lacombe, M.L.; Paggi, M.G. Nm23: Unraveling its biological function in cell differentiation. J. Cell. Physiol. 2000, 182, 144–149. [Google Scholar] [CrossRef]
- Boissan, M.; Dabernat, S.; Peuchant, E.; Schlattner, U.; Lascu, I.; Lacombe, M.L. The mammalian Nm23/NDPK family: From metastasis control to cilia movement. Mol. Cell. Biochem. 2009, 329, 51–62. [Google Scholar] [CrossRef]
- Palacios, F.; Schweitzer, J.K.; Boshans, R.L.; D’Souza-Schorey, C. ARF6-GTP recruits Nm23-H1 to facilitate dynamin-mediated endocytosis during adherens junctions disassembly. Nat. Cell Biol. 2002, 4, 929–936. [Google Scholar] [CrossRef]
- Boissan, M.; Montagnac, G.; Shen, Q.; Griparic, L.; Guitton, J.; Romao, M.; Sauvonnet, N.; Lagache, T.; Lascu, I.; Raposo, G.; et al. Membrane trafficking. Nucleoside diphosphate kinases fuel dynamin superfamily proteins with GTP for membrane remodeling. Science 2014, 344, 1510–1515. [Google Scholar] [CrossRef] [Green Version]
- Marino, N.; Marshall, J.C.; Collins, J.W.; Zhou, M.; Qian, Y.; Veenstra, T.; Steeg, P.S. Nm23-H1 binds to gelsolin and inactivates its actin-severing capacity to promote tumor cell motility and metastasis. Cancer Res. 2013, 73, 5949–5962. [Google Scholar] [CrossRef] [Green Version]
- Ignesti, M.; Barraco, M.; Nallamothu, G.; Woolworth, J.A.; Duchi, S.; Gargiulo, G.; Cavaliere, V.; Hsu, T. Notch signaling during development requires the function of awd, the Drosophila homolog of human metastasis suppressor gene Nm23. BMC Biol. 2014, 12, 12. [Google Scholar] [CrossRef] [Green Version]
- Thakur, R.K.; Yadav, V.K.; Kumar, A.; Singh, A.; Pal, K.; Hoeppner, L.; Saha, D.; Purohit, G.; Basundra, R.; Kar, A.; et al. Non-metastatic 2 (NME2)-mediated suppression of lung cancer metastasis involves transcriptional regulation of key cell adhesion factor vinculin. Nucleic Acids Res. 2014, 42, 11589–11600. [Google Scholar] [CrossRef] [Green Version]
- Moreno, V.; Gonzalo, P.; Gomez-Escudero, J.; Pollan, A.; Acin-Perez, R.; Breckenridge, M.; Yáñez-Mó, M.; Barreiro, O.; Orsenigo, F.; Kadomatsu, K.; et al. An EMMPRIN-gamma-catenin-Nm23 complex drives ATP production and actomyosin contractility at endothelial junctions. J. Cell Sci. 2014, 127, 3768–3781. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perina, D.; Bosnar, M.H.; Mikoc, A.; Muller, W.E.G.; Cetkovic, H. Characterization of Nme6-like gene/protein from marine sponge Suberites domuncula. Naunyn Schmied. Arch. Pharmacol. 2011, 384, 451–460. [Google Scholar] [CrossRef]
- Liu, P.; Choi, Y.K.; Qi, R.Z. NME7 is a functional component of the gamma-tubulin ring complex. Mol. Biol. Cell 2014, 25, 2017–2025. [Google Scholar] [CrossRef] [PubMed]
- Munier, A.; Feral, C.; Milon, L.; Pinon, V.P.; Gyapay, G.; Capeau, J.; Guellaen, G.; Lacombe, M.L. A new human Nm23 homologue (Nm23-H5) specifically expressed in testis germinal cells. FEBS Lett. 1998, 434, 289–294. [Google Scholar] [CrossRef] [Green Version]
- Munier, A.; Serres, C.; Kann, M.L.; Boissan, M.; Lesaffre, C.; Capeau, J.; Fouquet, J.P.; Lacombe, M.L. Nm23/NDP kinases in human male germ cells: Role in spermiogenesis and sperm motility? Exp. Cell Res. 2003, 289, 295–306. [Google Scholar] [CrossRef]
- Sarkar, H.; Arya, S.; Rai, U.; Majumdar, S.S. A Study of Differential Expression of Testicular Genes in Various Reproductive Phases of Hemidactylus flaviviridis (Wall Lizard) to Derive Their Association with Onset of Spermatogenesis and Its Relevance to Mammals. PLoS ONE 2016, 11, e0151150. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.J.; Cho, S.K.; Hwang, K.C.; Park, C.; Kim, J.H.; Park, S.B.; Hwang, S.; Kim, J.H. Nm23-M5 mediates round and elongated spermatid survival by regulating GPX-5 levels. FEBS Lett. 2009, 583, 1292–1298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ostrowski, L.E.; Blackburn, K.; Radde, K.M.; Moyer, M.B.; Schlatzer, D.M.; Moseley, A.; Boucher, R.C. A proteomic analysis of human cilia: Identification of novel components. Mol. Cell. Proteom. 2002, 1, 451–465. [Google Scholar] [CrossRef] [Green Version]
- Li, F.; Hu, G.; Jiang, Z.; Guo, J.; Wang, K.; Ouyang, K.; Wen, D.; Zhu, M.; Liang, J.; Qin, X.; et al. Identification of NME5 as a contributor to innate resistance to gemcitabine in pancreatic cancer cells. FEBS J. 2012, 279, 1261–1273. [Google Scholar] [CrossRef]
- Li, F.; Jiang, Z.; Wang, K.; Guo, J.; Hu, G.; Sun, L.; Wang, T.; Tang, X.; He, L.; Yao, J..; et al. Transactivation of the human NME5 gene by Sp1 in pancreatic cancer cells. Gene 2012, 503, 200–207. [Google Scholar] [CrossRef]
- Goc, A.; Kochuparambil, S.T.; Al-Husein, B.; Al-Azayzih, A.; Mohammad, S.; Somanath, P.R. Simultaneous modulation of the intrinsic and extrinsic pathways by simvastatin in mediating prostate cancer cell apoptosis. BMC Cancer 2012, 12, 409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ansoleaga, B.; Jove, M.; Schluter, A.; Garcia-Esparcia, P.; Moreno, J.; Pujol, A.; Pamplona, R.; Portero-Otín, M.; Ferrer, I. Deregulation of purine metabolism in Alzheimer’s disease. Neurobiol. Aging 2015, 36, 68–80. [Google Scholar] [CrossRef] [PubMed]
- Gopal, R.; Foster, K.W.; Yang, P. The DPY-30 domain and its flanking sequence mediate the assembly and modulation of flagellar radial spoke complexes. Mol. Cell. Biol. 2012, 32, 4012–4024. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsu, D.R.; Chuang, P.T.; Meyer, B.J. DPY-30, a nuclear protein essential early in embryogenesis for Caenorhabditis elegans dosage compensation. Development 1995, 121, 3323–3334. [Google Scholar]
- Jiang, H.; Shukla, A.; Wang, X.; Chen, W.Y.; Bernstein, B.E.; Roeder, R.G. Role for Dpy-30 in ES cell-fate specification by regulation of H3K4 methylation within bivalent domains. Cell 2011, 144, 513–525. [Google Scholar] [CrossRef] [Green Version]
- Mehus, J.G.; Deloukas, P.; Lambeth, D.O. NME6: A new member of the Nm23/nucleoside diphosphate kinase gene family located on human chromosome 3p21.3. Hum. Genet. 1999, 104, 454–459. [Google Scholar] [CrossRef]
- Edwards, L.; Gupta, R.; Filipp, F.V. Hypermutation of DPYD Deregulates Pyrimidine Metabolism and Promotes Malignant Progression. Mol. Cancer Res. 2016, 14, 196–206. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.H.; Ma, N.; Lin, Y.T.; Wu, C.C.; Hsiao, M.; Lu, F.L.; Yu, C.C.; Chen, S.Y.; Lu, J. A shRNA functional screen reveals Nme6 and Nme7 are crucial for embryonic stem cell renewal. Stem Cells 2012, 30, 2199–2211. [Google Scholar] [CrossRef]
- Carter, M.G.; Smagghe, B.J.; Stewart, A.K.; Rapley, J.A.; Lynch, E.; Bernier, K.J.; Keating, K.W.; Hatziioannou, V.M.; Hartman, E.J.; Bamdad, C.C. A Primitive Growth Factor, NME7, Is Sufficient to Induce Stable Naive State Human Pluripotency; Reprogramming in This Novel Growth Factor Confers Superior Differentiation. Stem Cells 2016, 34, 847–859. [Google Scholar] [CrossRef] [Green Version]
- Vogel, P.; Read, R.; Hansen, G.M.; Freay, L.C.; Zambrowicz, B.P.; Sands, A.T. Situs inversus in Dpcd/Poll−, Nme7− and Pkd1l1− mice. Vet. Pathol. 2010, 47, 120–131. [Google Scholar] [CrossRef]
- Ikeda, T. NDP kinase 7 is a conserved microtubule-binding protein preferentially expressed in ciliated cells. Cell Struct. Funct. 2010, 35, 23–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miranda, M.R.; de los Milagros Camara, M.; Bouvier, L.A.; Pereira, C.A. TcNDPK2, a Trypanosoma cruzi microtubule-associated nucleoside diphosphate kinase. Mol. Biochem. Parasitol. 2011, 177, 152–155. [Google Scholar] [CrossRef] [PubMed]
- Duriez, B.; Duquesnoy, P.; Escudier, E.; Bridoux, A.M.; Escalier, D.; Rayet, I.; Marcos, E.; Vojtek, A.M.; Bercher, J.F.; Amselem, S. A common variant in combination with a nonsense mutation in a member of the thioredoxin family causes primary ciliary dyskinesia. Proc. Natl. Acad. Sci. USA 2007, 104, 3336–3341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sadek, C.M.; Damdimopoulos, A.E.; Pelto-Huikko, M.; Gustafsson, J.A.; Spyrou, G.; Miranda-Vizuete, A. Sptrx-2, a fusion protein composed of one thioredoxin and three tandemly repeated NDP-kinase domains is expressed in human testis germ cells. Genes Cells 2011, 6, 1077–1090. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miranda-Vizuete, A.; Tsang, K.; Yu, Y.; Jimenez, A.; Pelto-Huikko, M.; Flickinger, C.J.; Sutovsky, P.; Oko, R. Cloning and developmental analysis of murid spermatid-specific thioredoxin-2 (SPTRX-2), a novel sperm fibrous sheath protein and autoantigen. J. Biol. Chem. 2003, 278, 44874–44885. [Google Scholar] [CrossRef] [Green Version]
- Sadek, C.M.; Jimenez, A.; Damdimopoulos, A.E.; Kieselbach, T.; Nord, M.; Gustafsson, J.A.; Spyrou, G.; Davis, E.C.; Oko, R.; van der Hoorn, F.A.; et al. Characterization of human thioredoxin-like 2—A novel microtubule-binding thioredoxin expressed predominantly in the cilia of lung airway epithelium and spermatid manchette and axoneme. J. Biol. Chem. 2003, 278, 13133–13142. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Yu, J.T.; Wang, H.F.; Hao, X.K.; Yang, Y.F.; Jiang, T.; Zhu, X.C.; Cao, L.; Zhang, D.Q.; Tan, L. Association between NME8 locus polymorphism and cognitive decline, cerebrospinal fluid and neuroimaging biomarkers in Alzheimer’s disease. PLoS ONE 2014, 9, e114777. [Google Scholar] [CrossRef] [Green Version]
- Chan, G.; White, C.C.; Winn, P.A.; Cimpean, M.; Replogle, J.M.; Glick, L.R.; Cuerdon, N.E.; Ryan, K.J.; Johnson, K.A.; Schneider, J.A.; et al. CD33 modulates TREM2: Convergence of Alzheimer loci. Nat. Neurosci. 2015, 18, 1556–1558. [Google Scholar] [CrossRef] [Green Version]
- Yoon, H.S.; Hackett, J.D.; Ciniglia, C.; Pinto, G.; Bhattacharya, D. A molecular timeline for the origin of photosynthetic eukaryotes. Mol. Biol. Evol. 2004, 21, 809–818. [Google Scholar] [CrossRef] [Green Version]
- Butterfield, N.J. Bangiomorpha pubescens n. gen., n. sp.: Implications for the evolution of sex, multicellularity, and the Mesoproterozoic/Neoproterozoic radiation of eukaryotes. Paleobiology 2000, 26, 386–404. [Google Scholar] [CrossRef]
- Collen, J.; Porcel, B.; Carre, W.; Ball, S.G.; Chaparro, C.; Tonon, T.; Barbeyron, T.; Michel, G.; Noel, B.; Valentin, K.; et al. Genome structure and metabolic features in the red seaweed Chondrus crispus shed light on evolution of the Archaeplastida. Proc. Natl. Acad. Sci. USA 2013, 110, 5247–5252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Milon, L.; Meyer, P.; Chiadmi, M.; Munier, A.; Johansson, M.; Karlsson, A.; Lascu, I.; Capeau, J.; Janin, J.; Lacombe, M.L. The human Nm23-H4 gene product is a mitochondrial nucleoside diphosphate kinase. J. Biol. Chem. 2000, 275, 14264–14272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perina, D.; Bosnar, M.H.; Bago, R.; Mikoc, A.; Harcet, M.; Deželjin, M.; Ćetkovć, H. Sponge non-metastatic Group I Nme gene/protein—Structure and function is conserved from sponges to humans. BMC Evol. Biol. 2011, 11, 87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bilitou, A.; Watson, J.; Gartner, A.; Ohnuma, S.I. The Nm23 family in development. Mol. Cell. Biochem. 2009, 329, 17–33. [Google Scholar] [CrossRef]
- Desvignes, T.; Pontarotti, P.; Bobe, J. Nme Gene Family Evolutionary History Reveals Pre-Metazoan Origins and High Conservation between Humans and the Sea Anemone, Nematostella vectensis. PLoS ONE 2010, 5, e15506. [Google Scholar] [CrossRef]
- Lascu, I.; Chaffotte, A.; Limbourg-Bouchon, B.; Veron, M. A Pro/Ser substitution in nucleoside diphosphate kinase of Drosophila melanogaster (mutation killer of prune) affects stability but not catalytic efficiency of the enzyme. J. Biol. Chem. 1992, 267, 12775–12781. [Google Scholar]
- Postel, E.H.; Weiss, V.H.; Beneken, J.; Kirtane, A. Mutational analysis of Nm23-H2/NDP kinase identifies the structural domains critical to recognition of a c-myc regulatory element. Proc. Natl. Acad. Sci. USA 1996, 93, 6892–6897. [Google Scholar] [CrossRef] [Green Version]
- Johansson, M.; Mackenzie-Hose, A.; Andersson, I.; Knorpp, C. Structure and mutational analysis of a plant mitochondrial nucleoside diphosphate kinase. Identification of residues involved in serine phosphorylation and oligomerization. Plant Physiol. 2004, 136, 3034–3042. [Google Scholar] [CrossRef] [Green Version]
- Postel, E.H. Cleavage of DNA by human Nm23-H2/nucleoside diphosphate kinase involves formation of a covalent protein-DNA complex. J. Biol. Chem. 1999, 274, 22821–22829. [Google Scholar] [CrossRef] [Green Version]
- Negroni, A.; Venturelli, D.; Tanno, B.; Amendola, R.; Ransac, S.; Cesi, V.; Calabretta, B.; Raschellà, G. Neuroblastoma specific effects of DR-Nm23 and its mutant forms on differentiation and apoptosis. Cell Death Differ. 2000, 7, 843–850. [Google Scholar] [CrossRef]
- McDermott, W.G.; Boissan, M.; Lacombe, M.L.; Steeg, P.S.; Horak, C.E. Nm23-H1 homologs suppress tumor cell motility and anchorage independent growth. Clin. Exp. Metastasis 2008, 25, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Dai, Z.; Xiao, W.; Jin, Y. Inhibition of Nm23-H1 gene expression in chronic myelogenous leukemia cells. Oncol. Lett. 2013, 6, 1093–1097. [Google Scholar] [CrossRef] [PubMed]
- Amendola, R.; Martinez, R.; Negroni, A.; Venturelli, D.; Tanno, B.; Calabretta, B.; Raschellà, G. DR-Nm23 expression affects neuroblastoma cell differentiation, integrin expression, and adhesion characteristics. Med. Pediatr. Oncol. 2001, 36, 93–96. [Google Scholar] [CrossRef]
- Lee, M.J.; Xu, D.Y.; Li, H.; Yu, G.R.; Leem, S.H.; Chu, I.S.; Kim, I.H.; Kim, D.G. Pro-oncogenic potential of Nm23-H2 in hepatocellular carcinoma. Exp. Mol. Med. 2012, 44, 214–224. [Google Scholar] [CrossRef] [PubMed]
- Almgren, M.A.; Henriksson, K.C.; Fujimoto, J.; Chang, C.L. Nucleoside diphosphate kinase A/Nm23-H1 promotes metastasis of NB69-derived human neuroblastoma. Mol. Cancer Res. 2004, 2, 387–394. [Google Scholar]
- Panayiotou, C.; Solaroli, N.; Karlsson, A. The many isoforms of human adenylate kinases. Int. J. Biochem. Cell Biol. 2014, 49, 75–83. [Google Scholar] [CrossRef]
- Van Rompay, A.R.; Johansson, M.; Karlsson, A. Identification of a novel human adenylate kinase. cDNA cloning, expression analysis, chromosome localization and characterization of the recombinant protein. Eur. J. Biochem. 1999, 261, 509–517. [Google Scholar] [CrossRef] [Green Version]
- Amiri, M.; Conserva, F.; Panayiotou, C.; Karlsson, A.; Solaroli, N. The human adenylate kinase 9 is a nucleoside mono- and diphosphate kinase. Int. J. Biochem. Cell Biol. 2013, 45, 925–931. [Google Scholar] [CrossRef]
- Lu, Q.; Inouye, M. Adenylate kinase complements nucleoside diphosphate kinase deficiency in nucleotide metabolism. Proc. Natl. Acad. Sci. USA 1996, 93, 5720–5725. [Google Scholar] [CrossRef] [Green Version]
- Wang, L. The role of Ureaplasma nucleoside monophosphate kinases in the synthesis of nucleoside triphosphates. FEBS J. 2007, 274, 1983–1990. [Google Scholar] [CrossRef]
- Nosaka, K.; Kawahara, M.; Masuda, M.; Satomi, Y.; Nishino, H. Association of nucleoside diphosphate kinase Nm23-H2 with human telomeres. Biochem. Biophys. Res. Commun. 1998, 243, 342–348. [Google Scholar] [CrossRef] [PubMed]
- Postel, E.H.; Berberich, S.J.; Rooney, J.W.; Kaetzel, D.M. Human Nm23/nucleoside diphosphate kinase regulates gene expression through DNA binding to nuclease-hypersensitive transcriptional elements. J. Bioenerg. Biomembr. 2000, 32, 277–284. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.I.; Park, S.; Jeoung, D.I.; Lee, H. Point mutations affecting the oligomeric structure of Nm23-H1 abrogates its inhibitory activity on colonization and invasion of prostate cancer cells. Biochem. Biophys. Res. Commun. 2003, 307, 281–289. [Google Scholar] [CrossRef]
- Dzeja, P.; Terzic, A. Adenylate kinase and AMP signaling networks: Metabolic monitoring, signal communication and body energy sensing. Int. J. Mol. Sci. 2009, 10, 1729–1772. [Google Scholar] [CrossRef] [Green Version]
- Wild, K.; Grafmuller, R.; Wagner, E.; Schulz, G.E. Structure, catalysis and supramolecular assembly of adenylate kinase from maize. Eur. J. Biochem. 1997, 250, 326–331. [Google Scholar] [CrossRef] [Green Version]
- Chi, S.; Wu, S.; Yu, J.; Wang, X.; Tang, X.; Liu, T. Phylogeny of C4-photosynthesis enzymes based on algal transcriptomic and genomic data supports an archaeal/proteobacterial origin and multiple duplication for most C4-related genes. PLoS ONE 2014, 9, e110154. [Google Scholar] [CrossRef]
- Xie, C.; Li, B.; Xu, Y.; Ji, D.; Chen, C. Characterization of the global transcriptome for Pyropia haitanensis (Bangiales, Rhodophyta) and development of cSSR markers. BMC Genom. 2013, 14, 107. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [Green Version]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [Green Version]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [Green Version]
- Abascal, F.; Zardoya, R.; Posada, D. ProtTest: Selection of best-fit models of protein evolution. Bioinformatics 2005, 21, 2104–2105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le, S.Q.; Gascuel, O. An improved general amino acid replacement matrix. Mol. Biol. Evol. 2008, 25, 1307–1320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agarwal, R.P.; Robison, B.; Parks, R.E., Jr. Nucleoside diphosphokinase from human erythrocytes. Methods Enzymol. 1978, 51, 376–386. [Google Scholar] [PubMed]
- Postel, E.H.; Ferrone, C.A. Nucleoside diphosphate kinase enzyme activity of Nm23-H2/PuF is not required for its DNA binding and in vitro transcriptional functions. J. Biol. Chem. 1994, 269, 8627–8630. [Google Scholar]






| Eukaryotic Supergroup | Organism | Nme Schematic Domain Presentation | ||
|---|---|---|---|---|
| Group I | Group II | |||
| Unikonts | Opistokonts | Homo sapiens |  |  |
| Spizellomyces punctatus |  |  | ||
| Apusozoa | Thecamonas trahens |  |  | |
| Amoebozoa | Dictyostelium purpureum |  |  | |
| Bikonts | Rhizaria | Bigelowiella natans |  |  |
| Chromalveolata | Phytophthora sojae |  |  | |
| Plants | Volvox carteri |  |  | |
| Chondrus crispus |  |  | ||
| Excavata | Naegleria gruberi |  |  | |
| Eukaryotic ancestor |  |  | ||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Perina, D.; Korolija, M.; Mikoč, A.; Halasz, M.; Herak Bosnar, M.; Ćetković, H. Characterization of Nme5-Like Gene/Protein from the Red Alga Chondrus Crispus. Mar. Drugs 2020, 18, 13. https://doi.org/10.3390/md18010013
Perina D, Korolija M, Mikoč A, Halasz M, Herak Bosnar M, Ćetković H. Characterization of Nme5-Like Gene/Protein from the Red Alga Chondrus Crispus. Marine Drugs. 2020; 18(1):13. https://doi.org/10.3390/md18010013
Chicago/Turabian StylePerina, Dragutin, Marina Korolija, Andreja Mikoč, Mirna Halasz, Maja Herak Bosnar, and Helena Ćetković. 2020. "Characterization of Nme5-Like Gene/Protein from the Red Alga Chondrus Crispus" Marine Drugs 18, no. 1: 13. https://doi.org/10.3390/md18010013
APA StylePerina, D., Korolija, M., Mikoč, A., Halasz, M., Herak Bosnar, M., & Ćetković, H. (2020). Characterization of Nme5-Like Gene/Protein from the Red Alga Chondrus Crispus. Marine Drugs, 18(1), 13. https://doi.org/10.3390/md18010013