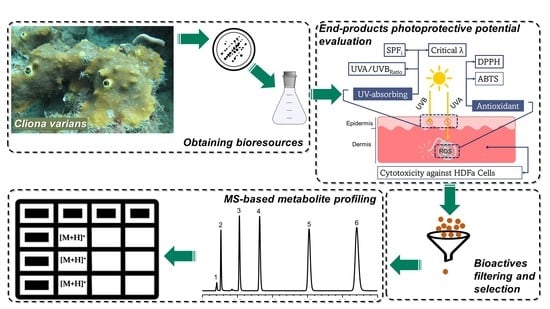
Cliona varians-Derived Actinomycetes as Bioresources of Photoprotection-Related Bioactive End-Products
Abstract
:1. Introduction
2. Results
2.1. Photoprotection-Related Activities of Actinobacterial Isolates
2.1.1. Radical Scavenging Capacity of Actinobacterial Isolates-Derived End-Products
2.1.2. UV-Absorbing Capacity of Actinobacterial Isolates-Derived Extracts
2.2. In Vitro Safety Evaluation of the Photoprotective Actinobacterial Extracts
2.3. Identification of Isolates with Photoprotective Potential by 16S rRNA Gene Sequencing
2.4. LC-MS-Based Characterization of Promising Microbial Extracts
3. Discussion
3.1. Actinomycetes as a Bioresource of Photoprotectants
3.2. Scavenging, UV-Absorbing, and Cytotoxicity Potential of Actinomycetes End-Products
3.3. Metabolite Profile of Promising Actinomycetes End-Products
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Sample Collection and Actinobacteria Isolation
4.3. Submerged Fermentation and Crude Extracts Preparation
4.4. Antioxidant Capacity Assays and Total Phenol and Flavonoid Content Measurements
4.4.1. DPPH and ABTS Radical Scavenging Assays
4.4.2. Total Phenol and Flavonoid Content
4.4.3. Determination of the In Vitro Sun Protection Factor and UV-Absorbing Profile
4.4.4. Sequencing of 16S rRNA Gene and Phylogenetic Analysis
4.4.5. Cytotoxicity Assay
4.4.6. Data Analysis
4.4.7. Metabolite Fingerprinting
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A

References
- Gilchrest, B.A. Actinic Injury. Annu. Rev. Med. 1990, 41, 199–210. [Google Scholar] [CrossRef]
- D’Orazio, J.; Jarrett, S.; Amaro-Ortiz, A.; Scott, T. UV radiation and the skin. Int. J. Mol. Sci. 2013, 14, 12222–12248. [Google Scholar] [CrossRef] [Green Version]
- Bolick, N.L.; Geller, A.C. Epidemiology of Melanoma. Hematol. Oncol. Clin. N. Am. 2021, 35, 57–72. [Google Scholar] [CrossRef]
- Parker, E.R. The influence of climate change on skin cancer incidence—A review of the evidence. Int. J. Womens Dermatol. 2021, 7, 17–27. [Google Scholar] [CrossRef]
- Krutmann, J.; Passeron, T.; Gilaberte, Y.; Granger, C.; Leone, G.; Narda, M.; Schalka, S.; Trullas, C.; Masson, P.; Lim, H.W. Photoprotection of the future: Challenges and opportunities. J. Eur. Acad. Dermatol. Venereol. 2020, 34, 447–454. [Google Scholar] [CrossRef]
- Heurung, A.R.; Raju, S.I.; Warshaw, E.M. Adverse reactions to sunscreen agents: Epidemiology, responsible irritants and allergens, clinical characteristics, and management. Dermatitis 2014, 25, 289–326. [Google Scholar] [CrossRef] [PubMed]
- Hiller, J.; Klotz, K.; Meyer, S.; Uter, W.; Hof, K.; Greiner, A.; Göen, T.; Drexler, H. Systemic availability of lipophilic organic UV filters through dermal sunscreen exposure. Environ. Int. 2019, 132, 105068. [Google Scholar] [CrossRef] [PubMed]
- Blasco, J.; Trombini, C.; Sendra, M.; Araujo, C.V.M. Environmental risk assessment of sunscreens. In Sunscreens in Coastal Ecosystems. Handbook of Environmental Chemistry; Springer International Publishing: Berlin, Germany, 2020; Volume 94, pp. 163–184. [Google Scholar]
- Schneider, S.L.; Lim, H.W. Review of environmental effects of oxybenzone and other sunscreen active ingredients. J. Am. Acad. Dermatol. 2019, 80, 266–271. [Google Scholar] [CrossRef] [PubMed]
- Mancuso, J.B.; Maruthi, R.; Wang, S.Q.; Lim, H.W. Sunscreens: An Update. Am. J. Clin. Dermatol. 2017, 18, 643–650. [Google Scholar] [CrossRef] [PubMed]
- Seebode, C.; Lehmann, J.; Emmert, S. Photocarcinogenesis and Skin Cancer Prevention Strategies: An Update. Anticancer Res. 2018, 38, 1371–1378. [Google Scholar] [CrossRef]
- Pinnell, S.R. Cutaneous photodamage, oxidative stress, and topical antioxidant protection. J. Am. Acad. Dermatol. 2003, 48, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Velasco, M.V.R.; Sauce, R.; de Oliveira, C.A.; de Oliveira Pinto, C.A.S.; Martinez, R.M.; Baah, S.; Almeida, T.S.; Rosado, C.; Baby, A.R. Active ingredients, mechanisms of action and efficacy tests of antipollution cosmetic and personal care products. Braz. J. Pharm. Sci. 2018, 54, e01003. [Google Scholar] [CrossRef]
- Cavinato, M.; Waltenberger, B.; Baraldo, G.; Grade, C.V.C.; Stuppner, H.; Jansen-Dürr, P. Plant extracts and natural compounds used against UVB-induced photoaging. Biogerontology 2017, 18, 499–516. [Google Scholar] [CrossRef]
- Atanasov, A.G.; Zotchev, S.B.; Dirsch, V.M.; Supuran, C.T. Natural products in drug discovery: Advances and opportunities. Nat. Rev. Drug Discov. 2021, 20, 200–216. [Google Scholar] [CrossRef] [PubMed]
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2019, 36, 122–173. [Google Scholar] [CrossRef] [Green Version]
- Sipkema, D.; Franssen, M.C.R.; Osinga, R.; Tramper, J.; Wijffels, R.H. Marine sponges as pharmacy. Mar. Biotechnol. 2005, 7, 142–162. [Google Scholar] [CrossRef]
- Esposito, R.; Ruocco, N.; Viel, T.; Federico, S.; Zupo, V.; Costantini, M. Sponges and their symbionts as a source of valuable compounds in cosmeceutical field. Mar. Drugs 2021, 19, 444. [Google Scholar] [CrossRef]
- Sacristán-Soriano, O.; Turon, X.; Hill, M. Microbiome structure of ecologically important bioeroding sponges (family Clionaidae): The role of host phylogeny and environmental plasticity. Coral Reefs 2020, 39, 1285–1298. [Google Scholar] [CrossRef]
- Ribes, M.; Dziallas, C.; Coma, R.; Riemann, L. Microbial diversity and putative diazotrophy in high- and low- microbial-abundance mediterranean sponges. Appl. Environ. Microbiol. 2015, 81, 5683–5693. [Google Scholar] [CrossRef] [Green Version]
- Eastgate, M.D.; Schmidt, M.A.; Fandrick, K.R. On the design of complex drug candidate syntheses in the pharmaceutical industry. Nat. Rev. Chem. 2017, 1, 1–16. [Google Scholar] [CrossRef]
- Atanasov, A.G.; Waltenberger, B.; Pferschy-Wenzig, E.-M.; Linder, T.; Wawrosch, C.; Uhrin, P.; Temml, V.; Wang, L.; Schwaiger, S.; Heiss, E.H.; et al. Discovery and resupply of pharmacologically active plant-derived natural products: A review. Biotechnol. Adv. 2015, 33, 1582–1614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molinski, T.F.; Dalisay, D.S.; Lievens, S.L.; Saludes, J.P. Drug development from marine natural products. Nat. Rev. Drug Discov. 2009, 8, 69–85. [Google Scholar] [CrossRef]
- Radjasa, O.K.; Vaske, Y.M.; Navarro, G.; Vervoort, H.C.; Tenney, K.; Linington, R.G.; Crews, P. Highlights of marine invertebrate-derived biosynthetic products: Their biomedical potential and possible production by microbial associants. Bioorg. Med. Chem. 2011, 19, 6658–6674. [Google Scholar] [CrossRef] [Green Version]
- Hug, J.J.; Krug, D.; Müller, R. Bacteria as genetically programmable producers of bioactive natural products. Nat. Rev. Chem. 2020, 4, 172–193. [Google Scholar] [CrossRef]
- Santos-Gandelman, J.; Giambiagi-deMarval, M.; Oelemann, W.; Laport, M. Biotechnological Potential of Sponge-Associated Bacteria. Curr. Pharm. Biotechnol. 2014, 15, 143–155. [Google Scholar] [CrossRef]
- Raimundo, I.; Silva, S.G.; Costa, R.; Keller-Costa, T. Bioactive secondary metabolites from octocoral-Associated microbes—New chances for blue growth. Mar. Drugs 2018, 16, 485. [Google Scholar] [CrossRef] [Green Version]
- Barka, E.A.; Vatsa, P.; Sanchez, L.; Gaveau-Vaillant, N.; Jacquard, C.; Meier-Kolthoff, J.P.; Klenk, H.-P.; Clément, C.; Ouhdouch, Y.; van Wezel, G.P. Taxonomy, Physiology, and Natural Products of Actinobacteria. Microbiol. Mol. Biol. Rev. 2016, 80, 1–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seipke, R.F.; Kaltenpoth, M.; Hutchings, M.I. Streptomyces as symbionts: An emerging and widespread theme? FEMS Microbiol. Rev. 2012, 36, 862–876. [Google Scholar] [CrossRef] [Green Version]
- Thomas, T.R.A.; Kavlekar, D.P.; LokaBharathi, P.A. Marine Drugs from Sponge-Microbe Association—A Review. Mar. Drugs 2010, 8, 1417–1468. [Google Scholar] [CrossRef] [Green Version]
- Poulsen, M.; Oh, D.-C.; Clardy, J.; Currie, C.R. Chemical Analyses of Wasp-Associated Streptomyces Bacteria Reveal a Prolific Potential for Natural Products Discovery. PLoS ONE 2011, 6, e16763. [Google Scholar] [CrossRef] [Green Version]
- Sánchez-Suárez, J.; Coy-Barrera, E.; Villamil, L.; Díaz, L. Streptomyces-Derived Metabolites with Potential Photoprotective Properties—A Systematic Literature Review and Meta-Analysis on the Reported Chemodiversity. Molecules 2020, 25, 3221. [Google Scholar] [CrossRef]
- Wang, S.Q.; Stanfield, J.W.; Osterwalder, U. In vitro assessments of UVA protection by popular sunscreens available in the United States. J. Am. Acad. Dermatol. 2008, 59, 934–942. [Google Scholar] [CrossRef] [PubMed]
- Ilyasov, I.R.; Beloborodov, V.L.; Selivanova, I.A.; Terekhov, R.P. ABTS/PP Decolorization Assay of Antioxidant Capacity Reaction Pathways. Int. J. Mol. Sci. 2020, 21, 1131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, D.; Boxin, O.U.; Prior, R.L. The chemistry behind antioxidant capacity assays. J. Agric. Food Chem. 2005, 53, 1841–1856. [Google Scholar] [CrossRef] [PubMed]
- Apak, R.; Gorinstein, S.; Böhm, V.; Schaich, K.M.; Özyürek, M.; Güçlü, K. Methods of measurement and evaluation of natural antioxidant capacity/activity (IUPAC Technical Report). Pure Appl. Chem. 2013, 85, 957–998. [Google Scholar] [CrossRef] [Green Version]
- Cole, C.; Shyr, T.; Ou-Yang, H. Metal oxide sunscreens protect skin by absorption, not by reflection or scattering. Photodermatol. Photoimmunol. Photomed. 2016, 32, 5–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarkany, R. Sun protection. Medicine (Baltimore) 2021, 49, 453–456. [Google Scholar] [CrossRef]
- Springsteen, A.; Yurek, R.; Frazier, M.; Carr, K.F. In vitro measurement of sun protection factor of sunscreens by diffuse transmittance. Anal. Chim. Acta 1999, 380, 155–164. [Google Scholar] [CrossRef]
- Paiva, J.P.; Diniz, R.R.; Leitão, A.C.; Cabral, L.M.; Fortunato, R.S.; Santos, B.A.M.C.; de Pádula, M. Insights and controversies on sunscreen safety. Crit. Rev. Toxicol. 2020, 50, 707–723. [Google Scholar] [CrossRef] [PubMed]
- Hughes, J.P.; Rees, S.S.; Kalindjian, S.B.; Philpott, K.L. Principles of early drug discovery. Br. J. Pharmacol. 2011, 162, 1239–1249. [Google Scholar] [CrossRef] [Green Version]
- Komaki, H. Reclassification of 15 Streptomyces species as synonyms of Streptomyces albogriseolus, Streptomyces althioticus, Streptomyces anthocyanicus, Streptomyces calvus, Streptomyces griseoincarnatus, Streptomyces mutabilis, Streptomyces pilosus or Streptomyces r. Int. J. Syst. Evol. Microbiol. 2019, 71, 004718. [Google Scholar] [CrossRef] [PubMed]
- Klimová, Z.; Hojerová, J.; Beránková, M. Skin absorption and human exposure estimation of three widely discussed UV filters in sunscreens—In vitro study mimicking real-life consumer habits. Food Chem. Toxicol. 2015, 83, 237–250. [Google Scholar] [CrossRef] [PubMed]
- Tampucci, S.; Burgalassi, S.; Chetoni, P.; Monti, D. Cutaneous Permeation and Penetration of Sunscreens: Formulation Strategies and In Vitro Methods. Cosmetics 2017, 5, 1. [Google Scholar] [CrossRef] [Green Version]
- Gilbert, E.; Pirot, F.; Bertholle, V.; Roussel, L.; Falson, F.; Padois, K. Commonly used UV filter toxicity on biological functions: Review of last decade studies. Int. J. Cosmet. Sci. 2013, 35, 208–219. [Google Scholar] [CrossRef]
- Klinubol, P.; Asawanonda, P.; Wanichwecharungruang, S.P. Transdermal Penetration of UV Filters. Skin Pharmacol. Physiol. 2008, 21, 23–29. [Google Scholar] [CrossRef]
- Benech-Kieffer, F.; Meuling, W.J.A.; Leclerc, C.; Roza, L.; Leclaire, J.; Nohynek, G. Percutaneous Absorption of Mexoryl SX® in Human Volunteers: Comparison with in vitro Data. Skin Pharmacol. Physiol. 2003, 16, 343–355. [Google Scholar] [CrossRef]
- Schymanski, E.L.; Jeon, J.; Gulde, R.; Fenner, K.; Ruff, M.; Singer, H.P.; Hollender, J. Identifying small molecules via high resolution mass spectrometry: Communicating confidence. Environ. Sci. Technol. 2014, 48, 2097–2098. [Google Scholar] [CrossRef]
- Rivera, G.S.M.; Beamish, C.R.; Wencewicz, T.A. Immobilized FhuD2 Siderophore-Binding Protein Enables Purification of Salmycin Sideromycins from Streptomyces violaceus DSM 8286. ACS Infect. Dis. 2018, 4, 845–859. [Google Scholar] [CrossRef]
- Kunihiro, S.; Kaneda, M. Glomecidin, a Novel Antifungal Cyclic Tetrapeptide Produced by Streptomyces lavendulae H698 SY2. J. Antibiot. (Tokyo) 2003, 56, 30–33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamdem, R.S.T.; Wang, H.; Wafo, P.; Ebrahim, W.; Özkaya, F.C.; Makhloufi, G.; Janiak, C.; Sureechatchaiyan, P.; Kassack, M.U.; Lin, W.; et al. Induction of new metabolites from the endophytic fungus Bionectria sp. through bacterial co-culture. Fitoterapia 2018, 124, 132–136. [Google Scholar] [CrossRef]
- Apsari, P.P.; Budiarti, S.; Wahyudi, A.T. Actinomycetes of rhizosphere soil producing antibacterial compounds against urinary tract infection bacteria. Biodiversitas 2019, 20, 1259–1265. [Google Scholar] [CrossRef] [Green Version]
- Krasnoff, S.B.; Englich, U.; Miller, P.G.; Shuler, M.L.; Glahn, R.P.; Donzelli, B.G.G.; Gibson, D.M. Metacridamides A and B, Macrocycles from Conidia of the Entomopathogenic Fungus Metarhizium acridum. J. Nat. Prod. 2012, 75, 175–180. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Zhang, D.; Zhang, M.; Liu, X.; Chen, R.; Zhao, J.; Li, L.; Wang, N.; Dai, J. Periconiasins I and J, two new cytochalasans from an endophytic fungus Periconia sp. Tetrahedron Lett. 2016, 57, 5794–5797. [Google Scholar] [CrossRef]
- Boros, C.; Smith, C.J.; Vasina, Y.; Che, Y.; Dix, A.B.; Darveaux, B.; Pearce, C. Isolation and Identification of the Icosalides—Cyclic Peptolides with Selective Antibiotic and Cytotoxic Activities. J. Antibiot. (Tokyo) 2006, 59, 486–494. [Google Scholar] [CrossRef] [PubMed]
- Yoshimura, A.; Covington, B.C.; Gallant, É.; Zhang, C.; Li, A.; Seyedsayamdost, M.R. Unlocking Cryptic Metabolites with Mass Spectrometry-Guided Transposon Mutant Selection. ACS Chem. Biol. 2020, 15, 2766–2774. [Google Scholar] [CrossRef] [PubMed]
- Jing, T.; Zhou, D.; Zhang, M.; Yun, T.; Qi, D.; Wei, Y.; Chen, Y.; Zang, X.; Wang, W.; Xie, J. Newly Isolated Streptomyces sp. JBS5-6 as a Potential Biocontrol Agent to Control Banana Fusarium Wilt: Genome Sequencing and Secondary Metabolite Cluster Profiles. Front. Microbiol. 2020, 11, 3036. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.-Y.; Hu, Y.-J.; Meng, F.-C.; Qu, S.-Y.; Wang, R.; Andersen, R.; Liao, Z.-H.; Chen, M. Bacillamidins A–G from a Marine-Derived Bacillus pumilus. Mar. Drugs 2018, 16, 326. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; MacMillan, J.B. Erythrazoles A–B, Cytotoxic Benzothiazoles from a Marine-Derived Erythrobacter sp. Org. Lett. 2011, 13, 6580–6583. [Google Scholar] [CrossRef] [Green Version]
- Korinth, G.; Wellner, T.; Schaller, K.H.; Drexler, H. Potential of the octanol–water partition coefficient (logP) to predict the dermal penetration behaviour of amphiphilic compounds in aqueous solutions. Toxicol. Lett. 2012, 215, 49–53. [Google Scholar] [CrossRef]
- Vinuchakkaravarthy, T.; Kumaravel, K.P.; Ravichandran, S.; Velmurugan, D. Active compound from the leaves of vitex negundo L. shows anti-inflammatory activity with evidence of inhibition for secretory phospholipase A2 through molecular docking. Bioinformation 2011, 7, 199–206. [Google Scholar] [CrossRef] [Green Version]
- Sander, T.; Freyss, J.; Von Korff, M.; Rufener, C. DataWarrior: An open-source program for chemistry aware data visualization and analysis. J. Chem. Inf. Model. 2015, 55, 460–473. [Google Scholar] [CrossRef]
- Cragg, G.M.; Newman, D.J. Natural products: A continuing source of novel drug leads. Biochim. Biophys. Acta—Gen. Subj. 2013, 1830, 3670–3695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haygood, M.G.; Schmidt, E.W.; Davidson, S.K.; Faulkner, D.J. Microbial symbionts of marine invertebrates: Opportunities for microbial biotechnology. J. Mol. Microbiol. Biotechnol. 1999, 1, 33–43. [Google Scholar] [PubMed]
- Omura, S.; Ikeda, H.; Ishikawa, J.; Hanamoto, A.; Takahashi, C.; Shinose, M.; Takahashi, Y.; Horikawa, H.; Nakazawa, H.; Osonoe, T.; et al. Genome sequence of an industrial microorganism Streptomyces avermitilis: Deducing the ability of producing secondary metabolites. Proc. Natl. Acad. Sci. USA 2001, 98, 12215–12220. [Google Scholar] [CrossRef] [Green Version]
- Seipke, R.F. Strain-level diversity of secondary metabolism in Streptomyces albus. PLoS ONE 2015, 10, e0116457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parte, A.C.; Sardà Carbasse, J.; Meier-Kolthoff, J.P.; Reimer, L.C.; Göker, M. List of Prokaryotic names with Standing in Nomenclature (LPSN) moves to the DSMZ. Int. J. Syst. Evol. Microbiol. 2020, 70, 5607–5612. [Google Scholar] [CrossRef]
- Jose, P.A.; Jebakumar, S.R.D. Non-streptomycete actinomycetes nourish the current microbial antibiotic drug discovery. Front. Microbiol. 2013, 4, 240. [Google Scholar] [CrossRef] [Green Version]
- Variyar, P.S.; Chander, R.; Venkatachalam, S.R.; Bongirwar, D.R. A new red pigment from an alkalophilic Micrococcus species. Indian J. Chem.—Sect. B Org. Med. Chem. 2002, 41, 232–233. [Google Scholar]
- Rostami, H.; Hamedi, H.; Yolmeh, M. Some biological activities of pigments extracted from Micrococcus roseus (PTCC 1411) and Rhodotorula glutinis (PTCC 5257). Int. J. Immunopathol. Pharmacol. 2016, 29, 684–695. [Google Scholar] [CrossRef] [Green Version]
- Karbalaei-Heidari, H.R.; Partovifar, M.; Memarpoor-Yazdi, M. Evaluation of the bioactive potential of secondary metabolites produced by a new marine Micrococcus species isolated from the Persian Gulf. Avicenna J. Med. Biotechnol. 2020, 12, 61–65. [Google Scholar]
- Vinoshna, S.; Akula, M.; Mishra, B. In vitro antioxidant efficacy of EPS obtained from Micrococcus luteus snist-CM 02: A brief study. J. Microbiol. Biotechnol. Food Sci. 2017, 6, 1199–1202. [Google Scholar] [CrossRef]
- Asker, M.M.S.; EL Sayed, O.H.; Mahmoud, M.G.; Ramadan, M.F. Chemical structure and antioxidant activity of a new exopolysaccharide produced from Micrococcus luteus. J. Genet. Eng. Biotechnol. 2014, 12, 121–126. [Google Scholar] [CrossRef] [Green Version]
- Tsao, S.-W.; Rudd, B.A.M.; He, X.-G.; Chang, C.-J.; Floss, H.G. Identification of a red pigment from Streptomyces coelicolor A3(2) as a mixture of prodigiosin derivatives. J. Antibiot. (Tokyo) 1985, 38, 128–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suryawanshi, R.K.; Patil, C.D.; Borase, H.P.; Narkhede, C.P.; Stevenson, A.; Hallsworth, J.E.; Patil, S.V. Towards an understanding of bacterial metabolites prodigiosin and violacein and their potential for use in commercial sunscreens. Int. J. Cosmet. Sci. 2015, 37, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Basílico, G.; Roger, C.A.; Seigelchifer, M.; Kerner, N. UV-specific DNA repair recombinant fusion enzyme: A new stable pharmacologically active principle suitable for photoprotection. J. Dermatol. Sci. 2005, 39, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Loh, W.L.C.; Huang, K.-C.; Ng, H.S.; Lan, J.C.-W. Exploring the fermentation characteristics of a newly isolated marine bacteria strain, Gordonia terrae TWRH01 for carotenoids production. J. Biosci. Bioeng. 2020, 130, 187–194. [Google Scholar] [CrossRef]
- Stahl, W.; Sies, H. Antioxidant activity of carotenoids. Mol. Aspects Med. 2003, 24, 345–351. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, F.; Banakar, S.; Li, Z. Comprehensive optimization of precursor-directed production of BC194 by Streptomyces rochei MB037 derived from the marine sponge Dysidea arenaria. Appl. Microbiol. Biotechnol. 2018, 102, 7865–7875. [Google Scholar] [CrossRef]
- Jiang, S.; Li, X.; Zhang, L.; Sun, W.; Dai, S.; Xie, L.; Liu, Y.; Lee, K.J. Culturable actinobacteria isolated from marine sponge Iotrochota sp. Mar. Biol. 2008, 153, 945–952. [Google Scholar] [CrossRef] [Green Version]
- Burke, K.E. Photoaging: The role of oxidative stress. G. Ital. Dermatol. Venereol. 2010, 145, 445–459. [Google Scholar]
- Munteanu, I.G.; Apetrei, C. Analytical Methods Used in Determining Antioxidant Activity: A Review. Int. J. Mol. Sci. 2021, 22, 3380. [Google Scholar] [CrossRef]
- Barclay, L.R.C.; Edwards, C.E.; Vinqvist, M.R. Media Effects on Antioxidant Activities of Phenols and Catechols. J. Am. Chem. Soc. 1999, 121, 6226–6231. [Google Scholar] [CrossRef]
- Oresajo, C.; Pillai, S.; Manco, M.; Yatskayer, M.; McDaniel, D. Antioxidants and the skin: Understanding formulation and efficacy. Dermatol. Ther. 2012, 25, 252–259. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.T.-H.; Chan, K.-G.; Chan, C.K.; Khan, T.M.; Lee, L.-H.; Goh, B.-H. Antioxidative Potential of a Streptomyces sp. MUM292 Isolated from Mangrove Soil. Biomed Res. Int. 2018, 2018, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, L.T.-H.; Chan, K.-G.; Khan, T.M.; Bukhari, S.I.; Saokaew, S.; Duangjai, A.; Pusparajah, P.; Lee, L.-H.; Goh, B.-H. Streptomyces sp. MUM212 as a Source of Antioxidants with Radical Scavenging and Metal Chelating Properties. Front. Pharmacol. 2017, 8, 276. [Google Scholar] [CrossRef]
- Tan, L.T.H.; Mahendra, C.K.; Yow, Y.Y.; Chan, K.G.; Khan, T.M.; Lee, L.H.; Goh, B.H. Streptomyces sp. MUM273b: A mangrove-derived potential source for antioxidant and UVB radiation protectants. Microbiologyopen 2019, 8, e859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaath, N.A. Ultraviolet filters. Photochem. Photobiol. Sci. 2010, 9, 464. [Google Scholar] [CrossRef] [PubMed]
- Mancebo, S.E.; Hu, J.Y.; Wang, S.Q. Sunscreens: A review of health benefits, regulations, and controversies. Dermatol. Clin. 2014, 32, 427–438. [Google Scholar] [CrossRef] [PubMed]
- DiNardo, J.C.; Downs, C.A. Dermatological and environmental toxicological impact of the sunscreen ingredient oxybenzone/benzophenone-3. J. Cosmet. Dermatol. 2018, 17, 15–19. [Google Scholar] [CrossRef]
- Fivenson, D.; Sabzevari, N.; Qiblawi, S.; Blitz, J.; Norton, B.B.; Norton, S.A. Sunscreens: UV filters to protect us: Part 2-Increasing awareness of UV filters and their potential toxicities to us and our environment. Int. J. Womens Dermatol. 2021, 7, 45–69. [Google Scholar] [CrossRef]
- Autier, P.; Boniol, M.; Severi, G.; Doré, J.F. Quantity of sunscreen used by European students. Br. J. Dermatol. 2001, 144, 288–291. [Google Scholar] [CrossRef]
- Bernerd, F.; Asselineau, D. UVA exposure of human skin reconstructed in vitro induces apoptosis of dermal fibroblasts: Subsequent connective tissue repair and implications in photoaging. Cell Death Differ. 1998, 5, 792–802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jackson, S.A.; Crossman, L.; Almeida, E.L.; Margassery, L.M.; Kennedy, J.; Dobson, A.D.W. Diverse and abundant secondary metabolism biosynthetic gene clusters in the genomes of marine sponge derived Streptomyces spp. Isolates. Mar. Drugs 2018, 16, 67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maipas, S.; Nicolopoulou-Stamati, P. Sun lotion chemicals as endocrine disruptors. Hormones 2015, 14, 32–46. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Ling, S.; Fang, Y.; Zhu, T.; Gu, Q.; Zhu, W.-M. Isolation, Structure Elucidation, and Antimycobacterial Properties of Dimeric Naphtho-γ-pyrones from the Marine-Derived FungusAspergillus carbonarius. Chem. Biodivers. 2008, 5, 93–100. [Google Scholar] [CrossRef]
- Wang, X.; Shaaban, K.A.; Elshahawi, S.I.; Ponomareva, L.V.; Sunkara, M.; Zhang, Y.; Copley, G.C.; Hower, J.C.; Morris, A.J.; Kharel, M.K.; et al. Frenolicins C-G, Pyranonaphthoquinones from Streptomyces sp. RM-4-15. J. Nat. Prod. 2013, 76, 1441–1447. [Google Scholar] [CrossRef] [Green Version]
- Cai, S.; Kong, X.; Wang, W.; Zhou, H.; Zhu, T.; Li, D.; Gu, Q. Aspergilazine A, a diketopiperazine dimer with a rare N-1 to C-6 linkage, from a marine-derived fungus Aspergillus taichungensis. Tetrahedron Lett. 2012, 53, 2615–2617. [Google Scholar] [CrossRef]
- Yu, H.; Li, S.-M. Two Cytochrome P450 Enzymes from Streptomyces sp. NRRL S-1868 Catalyze Distinct Dimerization of Tryptophan-Containing Cyclodipeptides. Org. Lett. 2019, 21, 7094–7098. [Google Scholar] [CrossRef]
- Kamdem, R.S.T.; Pascal, W.; Rehberg, N.; van Geelen, L.; Höfert, S.-P.; Knedel, T.-O.; Janiak, C.; Sureechatchaiyan, P.; Kassack, M.U.; Lin, W.; et al. Metabolites from the endophytic fungus Cylindrocarpon sp. isolated from tropical plant Sapium ellipticum. Fitoterapia 2018, 128, 175–179. [Google Scholar] [CrossRef]
- Hou, L.; Huang, H.; Li, H.; Wang, S.; Ju, J.; Li, W. Overexpression of a type III PKS gene affording novel violapyrones with enhanced anti-influenza A virus activity. Microb. Cell Fact. 2018, 17, 61. [Google Scholar] [CrossRef]
- Newton, G.G.F.; Abraham, E.P. Isolation of cephalosporin C, a penicillin-like antibiotic containing D-α-aminoadipic acid. Biochem. J. 1956, 62, 651–658. [Google Scholar] [CrossRef]
- Yu, H.; Serpe, E.; Romero, J.; Coque, J.-J.; Maeda, K.; Oelgeschlager, M.; Hintermann, G.; Liras, P.; Martin, J.-F.; Demain, A.L.; et al. Possible involvement of the lysine -aminotransferase gene (lat) in the expression of the genes encoding ACV synthetase (pcbAB) and isopenicillin N synthase (pcbC) in Streptomyces clavuligerus. Microbiology 1994, 140, 3367–3377. [Google Scholar] [CrossRef] [Green Version]
- Higgens, C.E.; Hamill, R.L.; Sands, T.H.; Hoehn, M.M.; Davis, N.E.; Nagarajan, R.; Boeck, L.D. The occurrence of deacetoxy-cephalosporin c in fungi and streptomycetes. J. Antibiot. (Tokyo) 1974, 27, 298–300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmitt, I.; Lumbsch, H.T. Ancient Horizontal Gene Transfer from Bacteria Enhances Biosynthetic Capabilities of Fungi. PLoS ONE 2009, 4, e4437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, D.K.; Pathak, J.; Pandey, A.; Singh, V.; Ahmed, H.; Rajneesh; Kumar, D.; Sinha, R.P. Ultraviolet-screening compound mycosporine-like amino acids in cyanobacteria: Biosynthesis, functions, and applications. In Advances in Cyanobacterial Biology; Elsevier: Amsterdam, The Netherlands, 2020; pp. 219–233. [Google Scholar]
- Edreva, A. The importance of non-photosynthetic pigments and cinnamic acid derivatives in photoprotection. Agric. Ecosyst. Environ. 2005, 106, 135–146. [Google Scholar] [CrossRef]
- Rouf, A.; Tanyeli, C. Bioactive thiazole and benzothiazole derivatives. Eur. J. Med. Chem. 2015, 97, 911–927. [Google Scholar] [CrossRef]
- Weber, T.; Charusanti, P.; Musiol-Kroll, E.M.; Jiang, X.; Tong, Y.; Kim, H.U.; Lee, S.Y. Metabolic engineering of antibiotic factories: New tools for antibiotic production in actinomycetes. Trends Biotechnol. 2015, 33, 15–26. [Google Scholar] [CrossRef]
- Wang, W.; Wang, H.; Hu, H.; Peng, H.; Zhang, X. Overexpression of afsR and Optimization of Metal Chloride to Improve Lomofungin Production in Streptomyces lomondensis S015. J. Microbiol. Biotechnol. 2015, 25, 672–680. [Google Scholar] [CrossRef]
- El-Housseiny, G.S.; Ibrahim, A.A.; Yassien, M.A.; Aboshanab, K.M. Production and statistical optimization of Paromomycin by Streptomyces rimosus NRRL 2455 in solid state fermentation. BMC Microbiol. 2021, 21, 34. [Google Scholar] [CrossRef]
- Zhu, S.; Duan, Y.; Huang, Y. The Application of Ribosome Engineering to Natural Product Discovery and Yield Improvement in Streptomyces. Antibiotics 2019, 8, 133. [Google Scholar] [CrossRef] [Green Version]
- Stankovic, N.; Radulovic, V.; Petkovic, M.; Vuckovic, I.; Jadranin, M.; Vasiljevic, B.; Nikodinovic-Runic, J. Streptomyces sp. JS520 produces exceptionally high quantities of undecylprodigiosin with antibacterial, antioxidative, and UV-protective properties. Appl. Microbiol. Biotechnol. 2012, 96, 1217–1231. [Google Scholar] [CrossRef] [PubMed]
- Matobole, R.M.; Van Zyl, L.J.; Parker-Nance, S.; Davies-Coleman, M.T.; Trindade, M. Antibacterial Activities of Bacteria Isolated from the Marine Sponges Isodictya compressa and Higginsia bidentifera Collected from Algoa Bay, South Africa. Mar. Drugs 2017, 15, 47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buitrago, D.; Buitrago-Villanueva, I.; Barbosa-Cornelio, R.; Coy-Barrera, E. Comparative examination of antioxidant capacity and fingerprinting of unfractionated extracts from different plant parts of quinoa (Chenopodium quinoa) grown under greenhouse conditions. Antioxidants 2019, 8, 238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Re, R.; Pellegrini, N.; Proteggente, A.; Pannala, A.; Yang, M.; Rice-Evans, C. Antioxidant activity applying an improved ABTS radical cation decolorization assay. Free Radic. Biol. Med. 1999, 26, 1231–1237. [Google Scholar] [CrossRef]
- Magalhães, L.M.; Santos, F.; Segundo, M.A.; Reis, S.; Lima, J.L.F.C. Rapid microplate high-throughput methodology for assessment of Folin-Ciocalteu reducing capacity. Talanta 2010, 83, 441–447. [Google Scholar] [CrossRef]
- Mansur, J.D.S.; Breder, M.N.R.; Mansur, M.C.D.A.; Azulay, R.D. Determinação do fator de proteção solar por espectrofotometria. An. Bras. Dermatol. 1986, 61, 121–124. [Google Scholar]
- Sayre, R.M.; Agin, P.P.; LeVee, G.J.; Marlowe, E. A comparison of in vivo and in vitro testing of sunscreening formulas. Photochem. Photobiol. 1979, 29, 559–566. [Google Scholar] [CrossRef]
- Morgulis, A.; Coulouris, G.; Raytselis, Y.; Madden, T.L.; Agarwala, R.; Schäffer, A.A. Database indexing for production MegaBLAST searches. Bioinformatics 2008, 24, 1757–1764. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Moumbock, A.F.A.; Gao, M.; Qaseem, A.; Li, J.; Kirchner, P.A.; Ndingkokhar, B.; Bekono, B.D.; Simoben, C.V.; Babiaka, S.B.; Malange, Y.I.; et al. StreptomeDB 3.0: An updated compendium of streptomycetes natural products. Nucleic Acids Res. 2021, 49, D600–D604. [Google Scholar] [CrossRef] [PubMed]
- Van Santen, J.A.; Poynton, E.F.; Iskakova, D.; McMann, E.; Alsup, T.A.; Clark, T.N.; Fergusson, C.H.; Fewer, D.P.; Hughes, A.H.; McCadden, C.A.; et al. The Natural Products Atlas 2.0: A database of microbially-derived natural products. Nucleic Acids Res. 2021, gkab941. [Google Scholar] [CrossRef] [PubMed]











| ID a | RT b | Molecular Formula | Adduct Type | m/z | Isomer Coincidence d | Identification Level e | ||
|---|---|---|---|---|---|---|---|---|
| Experimental | Calculated | Δ (ppm) c | ||||||
| 1 | 2.81 | C28H38N4O6 | [M+H]+ | 527.2838 | 527.2864 | 5.02 | 4 | 4 |
| 2 | 6.37 | C26H29NO5 | [M+2ACN+H] + | 518.2643 | 518.2650 | 1.39 | 7 | 4 |
| 3 | 14.50 | C22H26O6 | [M+H]+ | 387.1814 | 387.1802 | 2.96 | 13 | 4 |
| 4 | 15.22 | C17H30N2O3 | [M+2ACN+H] + | 393.2877 | 393.2860 | 4.22 | 1 | 3 |
| 5 | 15.69 | C26H28O6 | [M+H]+ | 437.1947 | 437.1959 | 2.66 | 5 | 4 |
| 6 | 16.30 | C24H30O6 | [M+H]+ | 415.2129 | 415.2115 | 3.35 | 9 | 4 |
| 7 | 20.00 | C37H55NO6 | [M+2ACN+H]+ | 692.4602 | 692.4633 | 4.47 | 1 | 3 |
| 8 | 20.12 | C32H54N4O7 | [M+ACN+H] + | 648.4338 | 648.4331 | 1.21 | 6 | 4 |
| 9 | 21.07 | C35H63NO3 | [M+H]+ | 546.4901 | 546.4881 | 3.69 | 3 | 4 |
| 10 | 21.79 | C42H75N7O17 | [M+ACN+H] + | 991.5576 | 991.5557 | 1.89 | 1 | 3 |
| 11 | 22.70 | C21H31NO2 | [M+2ACN+H] + | 412.2968 | 412.2959 | 2.22 | 1 | 3 |
| 12 | 22.85 | C27H37N7O7 | [M+2ACN+H] + | 654.3348 | 654.3358 | 1.48 | 1 | 3 |
| 13 | 22.93 | C31H42N2O7S | [M+2ACN+H] + | 669.3346 | 669.3317 | 4.32 | 1 | 3 |
| 14 | 23.02 | C42H63O4P | [M+H]+ | 663.4566 | 663.4537 | 4.38 | 1 | 3 |
| 15 | 23.34 | C34H60N4O10 | [M+H]+ | 685.4385 | 685.4382 | 0.48 | 1 | 3 |
| 16 | 23.44 | C22H38O2 | [M+ACN+H] + | 376.3202 | 376.321 | 2.19 | 3 | 4 |
| 17 | 25.29 | C32H26O10 | [M+ACN+H] + | 612.1849 | 612.1864 | 2.49 | 8 | 4 |
| ID a | Name | cLogP | Previously Described Bioactivities |
|---|---|---|---|
| 4 | Bacillamidin C | 3.191 | Antimicrobial, non-cytotoxic (evaluated against HepG2, A549, MDA-MB-231, SGC7901) [58]. |
| 7 | Metacridamide A | 8.213 | Cytotoxic against Caco-2, MCF-7, HepG2/C3A [53]. |
| 10 | Sideromycin A | −1.133 | Antimicrobial [49]. |
| 11 | Periconiasin J | 3.132 | Anti-HIV, non-cytotoxic (evaluated against MCF-7) [54]. |
| 12 | Glomecidin | −2.695 | Antifungal [50]. |
| 13 | Erythrazole A | 6.946 | Non-cytotoxic (evaluated against non-small cell lung cancer cell lines) [59]. |
| 14 | Tris(2,4-di-tert-butylphenyl)phosphate | 13.777 | Anti-inflammatory [61]. |
| 15 | Icosalide B | 2.551 | Antiviral and moderate cytotoxic activities (evaluated against MDCK cells) [55]. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sánchez-Suárez, J.; Villamil, L.; Coy-Barrera, E.; Díaz, L. Cliona varians-Derived Actinomycetes as Bioresources of Photoprotection-Related Bioactive End-Products. Mar. Drugs 2021, 19, 674. https://doi.org/10.3390/md19120674
Sánchez-Suárez J, Villamil L, Coy-Barrera E, Díaz L. Cliona varians-Derived Actinomycetes as Bioresources of Photoprotection-Related Bioactive End-Products. Marine Drugs. 2021; 19(12):674. https://doi.org/10.3390/md19120674
Chicago/Turabian StyleSánchez-Suárez, Jeysson, Luisa Villamil, Ericsson Coy-Barrera, and Luis Díaz. 2021. "Cliona varians-Derived Actinomycetes as Bioresources of Photoprotection-Related Bioactive End-Products" Marine Drugs 19, no. 12: 674. https://doi.org/10.3390/md19120674
APA StyleSánchez-Suárez, J., Villamil, L., Coy-Barrera, E., & Díaz, L. (2021). Cliona varians-Derived Actinomycetes as Bioresources of Photoprotection-Related Bioactive End-Products. Marine Drugs, 19(12), 674. https://doi.org/10.3390/md19120674