Okadaic Acid Exposure Induced Neural Tube Defects in Chicken (Gallus gallus) Embryos
Abstract
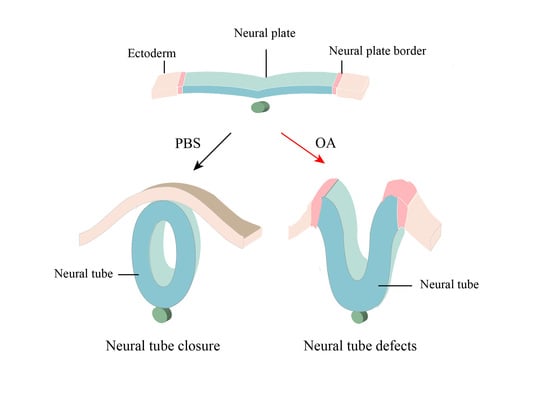
:1. Introduction
2. Results
2.1. OA Exposure Induced Craniofacial Abnormality in Early Stage Chick Embryos
2.2. OA Exposure Caused Craniofacial Abnormality in Late-stage Chick Embryos
2.3. OA Exposure Led to Abnormal Neurogenesis during Chick Embryo Development
2.4. OA Exposure Inhibited Cell Proliferation But Promoted Cell Apoptosis in the Developing Neural Tubes
2.5. OA Exposure Induce Oxidative Stress in Early Chick Embryo
2.6. Effects of OA on BMP/Shh Signaling Molecules
2.7. Transcriptome Analysis and qPCR Validation
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Ethical Statement
4.3. Chick Embryos
4.4. Immunofluorescent Staining
4.5. In Situ Hybridization
4.6. Fluorescent Microscopy
4.7. RNA Isolation and Quantitative Real-Time PCR Analysis
4.8. RNA-seq Assay
4.9. Western Blot
4.10. Detection of MDA Content and SOD Activity
4.11. Flow Cytometry Analysis
4.12. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Valdiglesias, V.; Prego-Faraldo, M.; Pásaro, E.; Méndez, J.; Laffon, B. Okadaic acid: More than a diarrheic toxin. Mar. Drugs 2013, 11, 4328–4349. [Google Scholar] [CrossRef] [Green Version]
- Reguera, B.; Riobó, P.; Rodríguez, F.; Díaz, P.; Pizarro, G.; Paz, B.; Franco, J.; Blanco, J. Dinophysis toxins: Causative organisms, distribution and fate in shellfish. Mar. Drugs 2014, 12, 394–461. [Google Scholar] [CrossRef] [PubMed]
- James, K.J.; Carey, B.; O’Halloran, J.; van Pelt, F.N.A.M.; Škrabáková, Z. Shellfish toxicity: Human health implications of marine algal toxins. Epidemiol. Infect. 2010, 138, 927–940. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vale, C.; Botana, L.M. Marine toxins and the cytoskeleton: Okadaic acid and dinophysistoxins. FEBS J. 2008, 275, 6060–6066. [Google Scholar] [CrossRef]
- Bialojan, C.; Takai, A. Inhibitory effect of a marine-sponge toxin, okadaic acid, on protein phosphatases. Specificity and kinetics. Biochem. J. 1988, 256, 283–290. [Google Scholar] [CrossRef] [PubMed]
- Holmes, F.B.; Luu, H.A.; Carrier, F.; Schmitz, F.J. Inhibition of protein phosphatases-1 and -2A with acanthifolicin: Comparison with diarrhetic shellfish toxins and identification of a region on okadaic acid important for phosphatase inhibition. FEBS Lett. 1990, 270, 216–218. [Google Scholar] [CrossRef] [Green Version]
- Valdiglesias, V.; Fernández-Tajes, J.; Costa, C.; Méndez, J.; Pásaro, E.; Laffon, B. Alterations in Metabolism-Related Genes Induced in SHSY5Y Cells by Okadaic Acid Exposure. J. Toxicol. Environ. Health Part A 2012, 75, 844–856. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.L.; Zhao, X.Y.; Ji, L.D.; Xu, J. Okadaic acid (OA): Toxicity, detection and detoxification. Toxicon 2019, 160, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Nuydens, R.; De Jong, M.; Van Den Kieboom, G.; Heers, C.; Dispersyn, G.; Cornelissen, F.; Nuyens, R.; Borgers, M.; Geerts, H. Okadaic acid-induced apoptosis in neuronal cells: Evidence for an abortive mitotic attempt. J. Neurochem. 1998, 70, 1124–1133. [Google Scholar] [CrossRef] [Green Version]
- Valdiglesias, V.; Laffon, B.; Pásaro, E.; Méndez, J. Okadaic acid induces morphological changes, apoptosis and cell cycle alterations in different human cell types. J. Environ. Monit. 2011, 13, 1831–1840. [Google Scholar] [CrossRef]
- Wang, R.; Li, J.; Zhao, Y.; Xing, Y.; Xue, X.; Zhang, J. Effects of okadaic acid combined with cisplatin on the proliferation and apoptosis of human lung adenocarcinoma A549 cells. Int. J. Clin. Exp. Med. 2017, 10, 10351–10358. [Google Scholar]
- Dietrich, J.; Schindler, M.; Lampen, A.; Braeuning, A.; Hessel-Pras, S. Comparison of long-term versus short-term effects of okadaic acid on the apoptotic status of human HepaRG cells. Chem.-Biol. Interact. 2020, 317, 108937. [Google Scholar] [CrossRef]
- Çakır, M.; Tekin, S.; Doğanyiğit, Z.; Erden, Y.; Soytürk, M.; Çiğremiş, Y.; Sandal, S. Cannabinoid type 2 receptor agonist JWH-133, attenuates Okadaic acid induced spatial memory impairment and neurodegeneration in rats. Life Sci. 2019, 217, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Chighladze, M.; Beselia, G.; Burjanadze, M.; Dashniani, M. Recognition memory impairment and neuronal degeneration induced by intracerebroventricular or intrahippocampal administration of okadaic acid. Eur. Neuropsychopharmacol. 2019, 29, S254–S255. [Google Scholar] [CrossRef]
- Kamat, P.K.; Rai, S.; Nath, C. Okadaic acid induced neurotoxicity: An emerging tool to study Alzheimer’s disease pathology. Neurotoxicology 2013, 37, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Koehler, D.; Williams, F.E. Utilizing zebrafish and okadaic acid to study Alzheimer’s disease. Neural Regen. Res. 2018, 13, 1538–1541. [Google Scholar] [PubMed]
- Casarini, L.; Franchini, A.; Malagoli, D.; Ottaviani, E. Evaluation of the effects of the marine toxin okadaic acid by using FETAX assay. Toxicol. Lett. 2007, 169, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Escoffier, N.; Gaudin, J.; Mezhoud, K.; Huet, H.; Chateau-Joubert, S.; Turquet, J.; Crespeau, F.; Edery, M. Toxicity to medaka fish embryo development of okadaic acid and crude extracts of Prorocentrum dinoflagellates. Toxicon 2007, 49, 1182–1192. [Google Scholar] [CrossRef]
- Jiao, Y.H.; Liu, M.; Wang, G.; Li, H.Y.; Liu, J.S.; Yang, X.X.; Yang, W.D. EMT is the major target for okadaic acid-suppressed the development of neural crest cells in chick embryo. Ecotoxicol. Environ. Saf. 2019, 180, 192–201. [Google Scholar] [CrossRef] [PubMed]
- Matias, W.; Creppy, E. Transplacental passage of [3H]-okadaic acid in pregnant mice measured by radioactivity and high-performance liquid chromatography. Hum. Exp. Toxicol. 1996, 15, 226–230. [Google Scholar] [CrossRef] [PubMed]
- Colas, J.F.; Schoenwolf, G.C. Towards a cellular and molecular understanding of neurulation. Dev. Dyn. 2001, 221, 117–145. [Google Scholar] [CrossRef] [PubMed]
- Copp, A.J.; Greene, N.D.; Murdoch, J.N. The genetic basis of mammalian neurulation. Nat. Rev. Genet. 2003, 4, 784. [Google Scholar] [CrossRef] [PubMed]
- Christianson, A.; Howson, C.P.; Modell, B. March of Dimes: Global Report on Birth Defects: The Hidden Toll of Dying and Disabled Children; March of Dimes Birth Defects Foundation: New York, NY, USA, 2005. [Google Scholar]
- Manning, S.M.; Jennings, R.; Madsen, J.R. Pathophysiology, prevention, and potential treatment of neural tube defects. Ment. Retard. Dev. Disabil. Res. Rev. 2000, 6, 6–14. [Google Scholar] [CrossRef]
- Padmanabhan, R. Etiology, pathogenesis and prevention of neural tube defects. Congenit. Anom. 2006, 46, 55–67. [Google Scholar] [CrossRef]
- Faez, T.; Skachkov, I.; Versluis, M.; Kooiman, K.; de Jong, N. In vivo characterization of ultrasound contrast agents: Microbubble spectroscopy in a chicken embryo. Ultrasound Med. Biol. 2012, 38, 1608–1617. [Google Scholar] [CrossRef]
- Lokman, N.A.; Elder, A.S.; Ricciardelli, C.; Oehler, M.K. Chick chorioallantoic membrane (CAM) assay as an in vivo model to study the effect of newly identified molecules on ovarian cancer invasion and metastasis. Int. J. Mol. Sci. 2012, 13, 9959–9970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Figueroa, D.; Signore, A.; Araneda, O.; Contreras, H.R.; Concha, M.; García, C. Toxicity and differential oxidative stress effects on zebrafish larvae following exposure to toxins from the okadaic acid group. J. Toxicol. Environ. Health Part A 2020, 83, 573–588. [Google Scholar] [CrossRef]
- Le Du, J.; Tovar-Ramírez, D.; Núñez-Vázquez, E. Embryotoxic effects of dissolved okadaic acid on the development of Longfin yellowtail Seriola rivoliana. Aquat. Toxicol. 2017, 190, 210–216. [Google Scholar] [CrossRef]
- McCarthy, M.; O’Halloran, J.; O’Brien, N.M.; van Pelt, F.F. Does the marine biotoxin okadaic acid cause DNA fragmentation in the blue mussel and the pacific oyster? Mar. Environ. Res. 2014, 101, 153–160. [Google Scholar] [CrossRef]
- Valdiglesias, V.; Laffon, B.; Pásaro, E.; Cemeli, E.; Anderson, D.; Méndez, J. Induction of oxidative DNA damage by the marine toxin okadaic acid depends on human cell type. Toxicon 2011, 57, 882–888. [Google Scholar] [CrossRef]
- Julien, J.P. Neurofilament functions in health and disease. Curr. Opin. Neurobiol. 1999, 9, 554–560. [Google Scholar] [CrossRef]
- Wang, H.; Wu, M.; Zhan, C.; Ma, E.; Yang, M.; Yang, X.; Li, Y. Neurofilament proteins in axonal regeneration and neurodegenerative diseases. Neural Regen. Res. 2012, 7, 620–626. [Google Scholar]
- Jouhilahti, E.M.; Peltonen, S.; Peltonen, J. Class III β-tubulin is a component of the mitotic spindle in multiple cell types. J. Histochem. Cytochem. 2008, 56, 1113–1119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lago, J.; Santaclara, F.; Vieites, J.M.; Cabado, A.G. Collapse of mitochondrial membrane potential and caspases activation are early events in okadaic acid-treated Caco-2 cells. Toxicon 2005, 46, 579–586. [Google Scholar] [CrossRef]
- Morana, S.J.; Wolf, C.M.; Li, J.; Reynolds, J.E.; Brown, M.K.; Eastman, A. The involvement of protein phosphatases in the activation of ICE/CED-3 protease, intracellular acidification, DNA digestion, and apoptosis. J. Biol. Chem. 1996, 271, 18263–18271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Härmälä-Braskén, A.S.; Mikhailov, A.; Söderström, T.S.; Meinander, A.; Holmström, T.H.; Damuni, Z.; Eriksson, J.E. Type-2A protein phosphatase activity is required to maintain death receptor responsiveness. Oncogene 2003, 22, 7677–7686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahn, K.H.; Kim, Y.S.; Kim, S.Y.; Huh, Y.; Park, C.; Jeong, J.W. Okadaic acid protects human neuroblastoma SH-SY5Y cells from 1-methyl-4-phenylpyridinium ion-induced apoptosis. Neurosci. Lett. 2009, 449, 93–97. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.; Horal, M.; Jain, S.; Wang, F.; Patel, R.; Loeken, M. Oxidant regulation of gene expression and neural tube development: Insights gained from diabetic pregnancy on molecular causes of neural tube defects. Diabetologia 2003, 46, 538–545. [Google Scholar] [CrossRef] [Green Version]
- Kensler, T.W.; Wakabayashi, N.; Biswal, S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu. Rev. Pharmacol. Toxicol. 2007, 47, 89–116. [Google Scholar] [CrossRef]
- Yamaguchi, Y.; Miura, M. How to form and close the brain: Insight into the mechanism of cranial neural tube closure in mammals. Cell. Mol. Life Sci. 2013, 70, 3171–3186. [Google Scholar] [CrossRef] [Green Version]
- Mahalik, S.K.; Vaze, D.; Lyngdoh, T.S.; Tewari, M.K.; Narasimhan, K.L. Embryogenesis of triple neural tube defects: Sonic hedgehog—A key? J. Clin. Pediatr. Surg. 2011, 46, e5–e8. [Google Scholar] [CrossRef]
- Jin, Y.M.; Wang, G.; Zhang, N.; Wei, Y.F.; Li, S.; Chen, Y.P.; Chuai, M.; Lee, H.S.S.; Hocher, B.; Yang, X. Changes in the osmolarity of the embryonic microenvironment induce neural tube defects. Mol. Reprod. Dev. 2015, 82, 365–376. [Google Scholar] [CrossRef] [PubMed]
- Dergunova, L.V.; Filippenkov, I.B.; Stavchansky, V.V.; Denisova, A.E.; Yuzhakov, V.V.; Mozerov, S.A.; Gubsky, L.V.; Limborska, S.A. Genome-wide transcriptome analysis using RNA-Seq reveals a large number of differentially expressed genes in a transient MCAO rat model. BMC Genom. 2018, 19, 655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawasaki, T.; Kawai, T. Toll-like receptor signaling pathways. Front. Immunol. 2014, 5, 461. [Google Scholar] [CrossRef] [Green Version]
- Curran, T.; Morgan, J.I. Fos: An immediate-early transcription factor in neurons. J. Neurobiol. 1995, 26, 403–412. [Google Scholar] [CrossRef]
- Velazquez, F.N.; Caputto, B.L.; Boussin, F.D. c-Fos importance for brain development. Aging 2015, 7, 1028–1029. [Google Scholar] [CrossRef] [Green Version]
- Van Dam, H.; Castellazzi, M. Distinct roles of Jun: Fos and Jun: ATF dimers in oncogenesis. Oncogene 2001, 20, 2453–2464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaulian, E.; Karin, M. AP-1 in cell proliferation and survival. Oncogene 2001, 20, 2390–2400. [Google Scholar] [CrossRef] [Green Version]
- Kovary, K.; Bravo, R. The jun and fos protein families are both required for cell cycle progression in fibroblasts. Mol. Cell. Biol. 1991, 11, 4466–4472. [Google Scholar] [CrossRef] [Green Version]
- Hsu, Y.J.; Hou, C.Y.; Lin, S.J.; Kuo, W.C.; Lin, H.T.; Lin, J.H.Y. The biofunction of orange-spotted grouper (Epinephelus coioides) CC chemokine ligand 4 (CCL4) in innate and adaptive immunity. Fish. Shellfish. Immunol. 2013, 35, 1891–1898. [Google Scholar] [CrossRef]
- Takahashi, T.; Kim, M.S.; Iwai-Shimada, M.; Fujimura, M.; Toyama, T.; Naganuma, A.; Hwang, G.W. Chemokine CCL4 induced in mouse brain has a protective role against methylmercury toxicity. Toxics 2018, 6, 36. [Google Scholar] [CrossRef] [Green Version]
- Regueiro, V.; Campos, M.; Morey, P.; Sauleda, J.; Agustí, A.; Garmendia, J.; Bengoechea, J. Lipopolysaccharide-binding protein and CD14 are increased in the bronchoalveolar lavage fluid of smokers. Eur. Respir. J. 2009, 33, 273–281. [Google Scholar] [CrossRef]
- Uesugi, T.; Froh, M.; Arteel, G.E.; Bradford, B.U.; Wheeler, M.D.; Gäbele, E.; Isayama, F.; Thurman, R.G. Role of lipopolysaccharide-binding protein in early alcohol-induced liver injury in mice. J. Immunol. 2002, 168, 2963–2969. [Google Scholar] [CrossRef] [Green Version]
- Pretorius, E.; Page, M.J.; Mbotwe, S.; Kell, D.B. Lipopolysaccharide-binding protein (LBP) can reverse the amyloid state of fibrin seen or induced in Parkinson’s disease. PLoS ONE 2018, 13, e0192121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baron, O.L.; Van West, P.; Industri, B.; Ponchet, M.; Dubreuil, G.; Gourbal, B.; Reichhart, J.M.; Coustau, C. Parental transfer of the antimicrobial protein LBP/BPI protects Biomphalaria glabrata eggs against oomycete infections. PLoS Pathog. 2013, 9, e1003792. [Google Scholar] [CrossRef] [Green Version]
- Henshel, D.S.; DeWitt, J.; Troutman, A. Using chicken embryos for teratology studies. Curr. Protoc. Toxicol. 2002, 14, 1–19. [Google Scholar] [CrossRef]
- Burt, D.W.; Bruley, C.; Dunn, I.C.; Jones, C.T.; Ramage, A.; Law, A.S.; Morrice, D.R.; Paton, I.R.; Smith, J.; Windsor, D.; et al. The dynamics of chromosome evolution in birds and mammals. Nature 1999, 402, 411–413. [Google Scholar] [CrossRef]
- Yaldiz, C.; Ceylan, D.; Sayin, M.; Kaçira, T.; Dilek, F.H. The effects of levetiracetam on neural tube development of chick embryos. Neurosurg. Q. 2015, 25, 468–471. [Google Scholar] [CrossRef]
- Ertekin, T.; Bilir, A.; Aslan, E.; Koca, B.; Turamanlar, O.; Ertekin, A.; Albay, S. The effect of diclofenac sodium on neural tube development in the early stage of chick embryos. Folia Morphol. 2019, 78, 307–313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chapman, S.C.; Collignon, J.; Schoenwolf, G.C.; Lumsden, A. Improved method for chick whole-embryo culture using a filter paper carrier. Dev. Dyn. 2001, 220, 284–289. [Google Scholar] [CrossRef]
- Henrique, D.; Adam, J.; Myat, A.; Chitnis, A.; Lewis, J.; Ish-Horowicz, D. Expression of a Delta homologue in prospective neurons in the chick. Nature 1995, 375, 787. [Google Scholar] [CrossRef]
- Jiao, Y.H.; Dou, M.; Wang, G.; Li, H.Y.; Liu, J.S.; Yang, X.X.; Yang, W.D. Exposure of okadaic acid alters the angiogenesis in developing chick embryos. Toxicon 2017, 133, 74–81. [Google Scholar] [CrossRef] [PubMed]
- Hellemans, J.; Mortier, G.; De Paepe, A.; Speleman, F.; Vandesompele, J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007, 8, R19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Feng, Z.; Wang, X.; Wang, X.; Zhang, X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010, 26, 136–138. [Google Scholar] [CrossRef]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [Green Version]
- Abdi, H. Bonferroni and Šidák corrections for multiple comparisons. In Encyclopedia of Measurement and Statistics; Sage: Thousand Oaks, CA, USA, 2007; pp. 103–107. [Google Scholar]








| ID | KO | Name | Product | Log2(FC) | Q-Value |
|---|---|---|---|---|---|
| 107056355 | K10030 | IL8 | interleukin 8 | −1.74 | 5.08 × 10−85 |
| 107056614 | K04398 | CASP8 | caspase 8 | 1.47 | 1.13 × 10−62 |
| 107057160 | K04430 | MAP2K4 | mitogen-activated protein kinase kinase 4 | 10.38 | 1.4 × 10−101 |
| 395196 | K04519 | IL1B | interleukin 1 beta | 3.86 | 1.76 × 10−8 |
| 395210 | K06250 | SPP1 | secreted phosphoprotein 1 | 1.03 | 0 |
| 395468 | K12964 | CCL4 | chemokine CC motif ligand 4 | 3.54 | 0 |
| 395872 | K10030 | IL8L1 | interleukin 8-like 1 | 7.46 | 2.33 × 10−20 |
| 396330 | K09447 | IRF7 | interferon regulatory factor 7 | 1.49 | 1.71 × 10−128 |
| 396512 | K04379 | FOS | proto-oncogene protein c-fos | 2.19 | 0 |
| 404671 | K05425 | IL12B | interleukin 12B | 1.77 | 7.7 × 10−20 |
| 416548 | K17783 | ERV1 | mitochondrial FAD-linked sulfhydryl oxidase | −1.01 | 2.29 × 10−19 |
| 424673 | K04448 | JUN | transcription factor AP-1 | 1.27 | 0 |
| 771461 | K05399 | LBP | lipopolysaccharide-binding protein | 1.37 | 2.1 × 10−10 |
| 395863 | K04097 | HPGDS | hematopoietic prostaglandin D synthase | 1.75 | 3.71 × 10−25 |
| Genes | Transcriptome Results Log2(FC) | qPCR Results |
|---|---|---|
| CASP8 | 1.47 | 1.16 |
| MAP2K4 | 10.38 | - |
| IL1B | 3.86 | - |
| SPP1 | 1.03 | - |
| CCL4 | 3.54 | - |
| FOS | 2.19 | 2.73 |
| IL12B | 1.77 | - |
| JUN | 1.27 | 1.94 |
| LBP | 1.37 | 1.51 |
| HPGDS | 1.75 | −0.62 |
| Genes | Primer Sequence 5′-3′ | Products Size (bp) |
|---|---|---|
| BMP4 | F: TTATAAAAGCTTGCGGCCGCAGAATATATGTTTGGCTGCGAAGGC R: GCTCTAGAAATTAACCCTCACTAAAGGGCGTGGTTGGTGGAGTTGAG | 860 |
| Shh | F: CCATCACTCCGAGGAATCGC R: AATTAACCCTCACTAAAGGGAGACCCAGCACATAGACACGTTG | 525 |
| Genes | Primer Sequence 5′-3′ | Products Size (bp) |
| RPL3 |
F: CTGGTGATGAAAAGCGGTAA R: CAAAGCAGGACAGTTGTTGG | 108 |
| RPS15 | F: TTCCGCAAGTTCACCTACAG | 165 |
| R: CAAAGCAGGACAGTTGTTGG | ||
| CASP8 |
F: TGGGAAAGTGGACAAGAGCC R: CATCTCTCCTTCACCAAGTAAGT | 73 |
| MAP2K4 |
F: GCATGCAGGGTAAACGCAAA R: AACCTTGCCGTGGACTTGAA | 70 |
| SPP1 |
F: GAGCGTAGAGAACGACAGCC R: CTCTAGCGTCTGGTTGCTGG | 139 |
| CCL4 |
F: AGCCTCCTCTGCCCCAG R: TCGCGCTCCTTCTTTGTGAT | 153 |
| FOS |
F: GCCGACATGATGTACCAGGG R: GACGGGTAGTAGGTGAGGCT | 101 |
| IL12B |
F: CACCAGCCGACTGAGATGTT R: GAGGTGGGTCTGGCTTTATGAT | 103 |
| LBP |
F: AAGGTTTGTGACAGCGTGGT R: ACGTTTGCTTCTGGCAAGGT | 77 |
| HPGDS |
F: GCCATTCCAACTGCATTCCC R: TTTTCTCCCTCTGCGAACCC | 84 |
| CREB |
F: AATGGATCTCTTGGGGCAGC R: ACCTGCCATTCCCATTTTTGT | 186 |
| CBP |
F: CCTCAACCACATGACGCACT R: GGCCGTCTTGAAGCTCATCTC | 111 |
| Nrf2 |
F: GGCCGTCTTGAAGCTCATCTC R: TGCCTCTCCTGCGTATATCTCG | 175 |
| Caspase3 |
F: CCACCGAGATACCGGACTGT R: GGAATGAGGACGAGCCAGAC | 173 |
| Caspase9 |
F: GGAATGAGGACGAGCCAGAC R: GTACCACGAGCCACTCACCTT | 119 |
| KEAP1 |
F: CTTCGCTGAGGTCTCCAAG R: CAGTCGTACTGCACCCAGTT | 142 |
| IL1B |
F: GGAGAGCAGCAGCCTCAG R: AGCCCTCCCATCCTTACCTT | 79 |
| KEAP1 | F: ACTTCGCTGAGGTCTCCAAG | 142 |
| R: CAGTCGTACTGCACCCAGTT | ||
| JUN | F: CCTCCCCTGTCCCCTATTGA | 99 |
| R: CCTTTTCCGGCATTTGGACG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiao, Y.; Wang, G.; Li, D.; Li, H.; Liu, J.; Yang, X.; Yang, W. Okadaic Acid Exposure Induced Neural Tube Defects in Chicken (Gallus gallus) Embryos. Mar. Drugs 2021, 19, 322. https://doi.org/10.3390/md19060322
Jiao Y, Wang G, Li D, Li H, Liu J, Yang X, Yang W. Okadaic Acid Exposure Induced Neural Tube Defects in Chicken (Gallus gallus) Embryos. Marine Drugs. 2021; 19(6):322. https://doi.org/10.3390/md19060322
Chicago/Turabian StyleJiao, Yuhu, Guang Wang, Dawei Li, Hongye Li, Jiesheng Liu, Xuesong Yang, and Weidong Yang. 2021. "Okadaic Acid Exposure Induced Neural Tube Defects in Chicken (Gallus gallus) Embryos" Marine Drugs 19, no. 6: 322. https://doi.org/10.3390/md19060322
APA StyleJiao, Y., Wang, G., Li, D., Li, H., Liu, J., Yang, X., & Yang, W. (2021). Okadaic Acid Exposure Induced Neural Tube Defects in Chicken (Gallus gallus) Embryos. Marine Drugs, 19(6), 322. https://doi.org/10.3390/md19060322