Identification of a New Antimicrobial, Desertomycin H, Utilizing a Modified Crowded Plate Technique
Abstract
:1. Introduction
2. Results
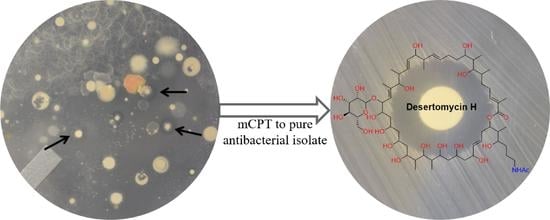
2.1. Waksman’s Crowded Plate Technique
2.2. Modified Crowded Plate Technique (mCPT)
2.3. d-Alanine Auxotrophs Amplify the Effects of Antibiotic Production and Simplify Purification
2.4. Dereplication Using Multidrug (MDR) and Extensively Drug-Resistant (XDR) Clinical Isolates
2.5. Dereplication Using the Antibiotic Resistance Platform (ARP)
2.6. Chemical Analysis of Mixed Fermentation Cultures
2.7. Purification and Identification of Antimicrobial Entities
2.8. Antibacterial Evaluation
3. Discussion
4. Materials and Methods
4.1. General Experimental Details
4.2. Bacterial Strains
4.3. Reagents, Soil Samples, and Culture Conditions
4.4. Crowded Plate Technique
4.5. Modified Crowded Plate Technique
4.6. Spread-Patch Assays
4.7. 16S rRNA Gene Sequencing and Analysis
4.8. Chemical Extractions and Testing
4.9. Activity-Guided Fractionation, Purification, and Structural Analysis
4.10. Antibacterial Assay
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Critically Important Antimicrobials for Human Medicine; World Health Organization: Geneva, Switzerland, 2019. [Google Scholar]
- World Health Organization. Critically Important Antimicrobials for Human Medicine: Ranking of Antimicrobial Agents for Risk Management of Antimicrobial Resistance Due to Non-Human Use; World Health Organization: Geneva, Switzerland, 2017. [Google Scholar]
- Ventola, C.L. The antibiotic resistance crisis: Part 1: Causes and threats. Pharm. Ther. 2015, 40, 277. [Google Scholar]
- Centers for Disease Control and Prevention; US Department of Health and Human Services; Centers for Disease Control and Prevention. Antibiotic Resistance Threats in the United States; US Department of Health and Human Services: Atlanta, GA, USA, 2013.
- Cassini, A.; Högberg, L.D.; Plachouras, D.; Quattrocchi, A.; Hoxha, A.; Simonsen, G.S.; Colomb-Cotinat, M.; Kretzschmar, M.E.; Devleesschauwer, B.; Cecchini, M. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: A population-level modelling analysis. Lancet Infect. Dis. 2019, 19, 56–66. [Google Scholar] [CrossRef] [Green Version]
- Holdren, J.; Lander, E.; Press, W. Report to the President on Combatting Antibiotic Resistance. Pres. Counc. Advis. Sci. Technol. 2014, 165. [Google Scholar]
- Thorpe, K.E.; Joski, P.; Johnston, K.J. Antibiotic-resistant infection treatment costs have doubled since 2002, now exceeding $2 billion annually. Health Aff. 2018, 37, 662–669. [Google Scholar] [CrossRef]
- O’neill, J. Antimicrobial resistance. In Tackling a Crisis for the Health and Wealth of Nations; Review on Anitmicrobial Resistance: London, UK, 2014. [Google Scholar]
- Fernandes, P.; Martens, E. Antibiotics in late clinical development. Biochem. Pharmacol. 2017, 133, 152–163. [Google Scholar] [CrossRef] [Green Version]
- Shore, C.K.; Coukell, A. Roadmap for antibiotic discovery. Nat. Microbiol. 2016, 1, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Kealey, C.; Creaven, C.; Murphy, C.; Brady, C. New approaches to antibiotic discovery. Biotechnol. Lett. 2017, 39, 805–817. [Google Scholar] [CrossRef] [PubMed]
- Bentley, S.D.; Chater, K.F.; Cerdeño-Tárraga, A.-M.; Challis, G.L.; Thomson, N.; James, K.D.; Harris, D.E.; Quail, M.A.; Kieser, H.; Harper, D. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3 (2). Nature 2002, 417, 141–147. [Google Scholar] [CrossRef]
- Ling, L.L.; Schneider, T.; Peoples, A.J.; Spoering, A.L.; Engels, I.; Conlon, B.P.; Mueller, A.; Schäberle, T.F.; Hughes, D.E.; Epstein, S. A new antibiotic kills pathogens without detectable resistance. Nature 2015, 517, 455–459. [Google Scholar] [CrossRef] [PubMed]
- Hover, B.M.; Kim, S.-H.; Katz, M.; Charlop-Powers, Z.; Owen, J.G.; Ternei, M.A.; Maniko, J.; Estrela, A.B.; Molina, H.; Park, S. Culture-independent discovery of the malacidins as calcium-dependent antibiotics with activity against multidrug-resistant Gram-positive pathogens. Nat. Microbiol. 2018, 3, 415–422. [Google Scholar] [CrossRef] [Green Version]
- Van der Meij, A.; Worsley, S.F.; Hutchings, M.I.; Van Wezel, G.P. Chemical ecology of antibiotic production by actinomycetes. FEMS Microbiol. Rev. 2017, 41, 392–416. [Google Scholar] [CrossRef]
- Abrudan, M.I.; Smakman, F.; Grimbergen, A.J.; Westhoff, S.; Miller, E.L.; Van Wezel, G.P.; Rozen, D.E. Socially mediated induction and suppression of antibiosis during bacterial coexistence. Proc. Natl. Acad. Sci. USA 2015, 112, 11054–11059. [Google Scholar] [CrossRef] [Green Version]
- Debois, D.; Fernandez, O.; Franzil, L.; Jourdan, E.; De Brogniez, A.; Willems, L.; Clément, C.; Dorey, S.; De Pauw, E.; Ongena, M. Plant polysaccharides initiate underground crosstalk with bacilli by inducing synthesis of the immunogenic lipopeptide surfactin. Environ. Microbiol. Rep. 2015, 7, 570–582. [Google Scholar] [CrossRef]
- Rigali, S.; Titgemeyer, F.; Barends, S.; Mulder, S.; Thomae, A.W.; Hopwood, D.A.; Van Wezel, G.P. Feast or famine: The global regulator DasR links nutrient stress to antibiotic production by Streptomyces. EMBO Rep. 2008, 9, 670–675. [Google Scholar] [CrossRef] [Green Version]
- Seyedsayamdost, M.R. High-throughput platform for the discovery of elicitors of silent bacterial gene clusters. Proc. Natl. Acad. Sci. USA 2014, 111, 7266–7271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Onaka, H.; Mori, Y.; Igarashi, Y.; Furumai, T. Mycolic acid-containing bacteria induce natural-product biosynthesis in Streptomyces species. Appl. Environ. Microbiol. 2011, 77, 400–406. [Google Scholar] [CrossRef] [Green Version]
- Mohamed, O.G.; Khalil, Z.G.; Salim, A.A.; Cui, H.; Blumenthal, A.; Capon, R.J. Lincolnenins A–D: Isomeric Bactericidal Bianthracenes from Streptomyces lincolnensis. J. Org. Chem. 2020. [Google Scholar] [CrossRef]
- Traxler, M.F.; Watrous, J.D.; Alexandrov, T.; Dorrestein, P.C.; Kolter, R. Interspecies interactions stimulate diversification of the Streptomyces coelicolor secreted metabolome. MBio 2013, 4, e00459-13. [Google Scholar] [CrossRef] [Green Version]
- Helfrich, E.J.; Vogel, C.M.; Ueoka, R.; Schäfer, M.; Ryffel, F.; Müller, D.B.; Probst, S.; Kreuzer, M.; Piel, J.; Vorholt, J.A. Bipartite interactions, antibiotic production and biosynthetic potential of the Arabidopsis leaf microbiome. Nat. Microbiol. 2018, 3, 909–919. [Google Scholar] [CrossRef]
- Waksman, S. Microbial Antagonism and Antibiotic Substance; The Commonwealth Fund: New York, NY, USA, 1945. [Google Scholar]
- Wilska, A. Spray inoculation of plates in the detection of antagonistic microorganisms. Microbiology 1947, 1, 368–370. [Google Scholar]
- Stansly, P. A bacterial spray apparatus useful in searching for antibiotic-producing microorganisms. J. Bacteriol. 1947, 54, 443. [Google Scholar] [CrossRef] [Green Version]
- Foster, J.W.; Woodruff, H.B. Bacillin, a new antibiotic substance from a soil isolate of Bacillus subtilis. J. Bacteriol. 1946, 51, 363. [Google Scholar] [CrossRef] [Green Version]
- Kelner, A. A method for investigating large microbial populations for antibiotic activity. J. Bacteriol. 1948, 56, 157. [Google Scholar] [CrossRef] [Green Version]
- Meyers, E.; Smith, D.A.; Donovick, R. Bioautographic technique for a rapid survey of microbial populations for the production of antibiotics and other metabolites. Appl. Microbiol. 1968, 16, 10–12. [Google Scholar] [CrossRef]
- Waksman, S.A.; Lechevalier, H.A. The Actinomycetes. Vol. III. Antibiotics of Actinomycetes. J. Pharm. Sci. 1962, 51, 808. [Google Scholar]
- Hurley, A.; Chevrette, M.G.; Acharya, D.D.; Lozano, G.L.; Garavito, M.; Heinritz, J.; Balderrama, L.; Beebe, M.; DenHartog, M.L.; Corinaldi, K. Tiny Earth: A Big Idea for STEM Education and Antibiotic Discovery. mBio 2021, 12. [Google Scholar] [CrossRef]
- Painter, M.M.; Zimmerman, G.E.; Merlino, M.S.; Robertson, A.W.; Terry, V.H.; Ren, X.; McLeod, M.R.; Gomez-Rodriguez, L.; Garcia, K.A.; Leonard, J.A. Concanamycin A counteracts HIV-1 Nef to enhance immune clearance of infected primary cells by cytotoxic T lymphocytes. Proc. Natl. Acad. Sci. USA 2020, 117, 23835–23846. [Google Scholar] [CrossRef]
- Robertson, A.W.; Sandoval, J.; Mohamed, O.G.; Zhuang, Y.; Gallagher, E.E.; Schmidt, J.; Caratelli, L.; Menon, A.; Schultz, P.J.; Torrez, R.M. Discovery of surfactins as inhibitors of microRNA processing using cat-ELCCA. ACS Med. Chem. Lett. 2021. [Google Scholar] [CrossRef]
- Marcolefas, E.; Leung, T.; Okshevsky, M.; McKay, G.; Hignett, E.; Hamel, J.; Aguirre, G.; Blenner-Hassett, O.; Boyle, B.; Lévesque, R.C. Culture-dependent bioprospecting of bacterial isolates from the Canadian high Arctic displaying antibacterial activity. Front. Microbiol. 2019, 10, 1836. [Google Scholar] [CrossRef] [Green Version]
- Cox, G.; Sieron, A.; King, A.M.; De Pascale, G.; Pawlowski, A.C.; Koteva, K.; Wright, G.D. A common platform for antibiotic dereplication and adjuvant discovery. Cell Chem. Biol. 2017, 24, 98–109. [Google Scholar] [CrossRef] [Green Version]
- Lewis, K. Platforms for antibiotic discovery. Nat. Rev. Drug Discov. 2013, 12, 371–387. [Google Scholar] [CrossRef]
- Robbins, N.; Spitzer, M.; Wang, W.; Waglechner, N.; Patel, D.J.; O’Brien, J.S.; Ejim, L.; Ejim, O.; Tyers, M.; Wright, G.D. Discovery of ibomycin, a complex macrolactone that exerts antifungal activity by impeding endocytic trafficking and membrane function. Cell Chem. Biol. 2016, 23, 1383–1394. [Google Scholar] [CrossRef] [Green Version]
- Behie, S.W.; Bonet, B.; Zacharia, V.M.; McClung, D.J.; Traxler, M.F. Molecules to ecosystems: Actinomycete natural products in situ. Front. Microbiol. 2017, 7, 2149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yim, G.; Huimi Wang, H.; Davies Frs, J. Antibiotics as signalling molecules. Philos. Trans. R. Soc. B Biol. Sci. 2007, 362, 1195–1200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaz Jauri, P.; Kinkel, L.L. Nutrient overlap, genetic relatedness and spatial origin influence interaction-mediated shifts in inhibitory phenotype among Streptomyces spp. FEMS Microbiol. Ecol. 2014, 90, 264–275. [Google Scholar] [CrossRef] [Green Version]
- Pishchany, G.; Mevers, E.; Ndousse-Fetter, S.; Horvath, D.J.; Paludo, C.R.; Silva-Junior, E.A.; Koren, S.; Skaar, E.P.; Clardy, J.; Kolter, R. Amycomicin is a potent and specific antibiotic discovered with a targeted interaction screen. Proc. Natl. Acad. Sci. USA 2018, 115, 10124–10129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scherlach, K.; Graupner, K.; Hertweck, C. Molecular bacteria-fungi interactions: Effects on environment, food, and medicine. Annu. Rev. Microbiol. 2013, 67, 375–397. [Google Scholar] [CrossRef]
- Uri, J.; Bognar, R.; Bekesi, I.; Varga, B. Desertomycin, a new crystalline antibiotic with antibacterial and cytostatic action. Nature 1958, 182, 401. [Google Scholar] [CrossRef]
- Ivanova, V. New Macrolactone of the Desertomycin Family from Streptomyces spectabilis. Prep. Biochem. Biotechnol. 1997, 27, 19–38. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Lee, J.; Tezuka, K.; Kishi, Y. Toward creation of a universal NMR database for the stereochemical assignment of acyclic compounds: The case of two contiguous propionate units. Org. Lett. 1999, 1, 2177–2180. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Tan, C.-H.; Kishi, Y. Toward creation of a universal NMR database for stereochemical assignment: Complete structure of the desertomycin/oasomycin class of natural products. J. Am. Chem. Soc. 2001, 123, 2076–2078. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Kobayashi, Y.; Tezuka, K.; Kishi, Y. Toward creation of a universal NMR database for the stereochemical assignment of acyclic compounds: Proof of concept. Org. Lett. 1999, 1, 2181–2184. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.H.; Kobayashi, Y.; Kishi, Y. Stereochemical assignment of the C21–C38 portion of the desertomycin/oasomycin class of natural products by using universal NMR databases: Proof. Angew. Chem. 2000, 112, 4452–4454. [Google Scholar] [CrossRef]
- Leisner, J.; Haaber, J. Intraguild predation provides a selection mechanism for bacterial antagonistic compounds. Proc. R. Soc. B Biol. Sci. 2012, 279, 4513–4521. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Shamaei-Tousi, A.; Liu, Y.; Coates, A. A new approach for the discovery of antibiotics by targeting non-multiplying bacteria: A novel topical antibiotic for staphylococcal infections. PLoS ONE 2010, 5, e11818. [Google Scholar] [CrossRef] [PubMed]
- Seyedsayamdost, M.R.; Traxler, M.F.; Clardy, J.; Kolter, R. Old meets new: Using interspecies interactions to detect secondary metabolite production in actinomycetes. Methods Enzymol. 2012, 517, 89–109. [Google Scholar]
- Nothias, L.-F.; Petras, D.; Schmid, R.; Dührkop, K.; Rainer, J.; Sarvepalli, A.; Protsyuk, I.; Ernst, M.; Tsugawa, H.; Fleischauer, M. Feature-based molecular networking in the GNPS analysis environment. Nat. Methods 2020, 17, 905–908. [Google Scholar] [CrossRef]
- Lazazzera, B.A.; Solomon, J.M.; Grossman, A.D. An exported peptide functions intracellularly to contribute to cell density signaling in B. subtilis. Cell 1997, 89, 917–925. [Google Scholar] [CrossRef] [Green Version]
- Strych, U.; Huang, H.-C.; Krause, K.L.; Benedik, M.J. Characterization of the alanine racemases from Pseudomonas aeruginosa PAO1. Curr. Microbiol. 2000, 41, 290–294. [Google Scholar] [CrossRef]





| Media + Coculture | Target Organism | ||
|---|---|---|---|
| Strain | S. aureus | VISA | |
| PAP57 | A3M | 8 | 7 |
| A3M-Corny | 9 | 8 | |
| A3M-Myco | 8 | 8 | |
| A3M-Rhodo | 9 | 8 | |
| PAP58 | A3M | 15 | 18 |
| A3M-Corny | 16 | 18 | |
| A3M-Myco | 12 | 10 | |
| A3M-Rhodo | 13 | 11 | |
| PAP60 | A3M | 15 | 12 |
| A3M-Corny | 13 | 10 | |
| A3M-Myco | 13 | 12 | |
| A3M-Rhodo | 13 | 13 | |
| PAP61 | A3M | 9 | 6 |
| A3M-Corny | 9 | 11 | |
| A3M-Myco | 10 | 12 | |
| A3M-Rhodo | 8 | 11 | |
| PAP62 | A3M | 10 | 9 |
| A3M-Corny | 13 | 14 | |
| A3M-Myco | 12 | 13 | |
| A3M-Rhodo | 14 | 15 | |
| PAP203 | A3M | 10 | 9 |
| A3M-Corny | 11 | 10 | |
| A3M-Myco | 10 | 9 | |
| A3M-Rhodo | 10 | 8 | |
| Pos. | 1 | 2 | Pos. | 1 | 2 | ||||
|---|---|---|---|---|---|---|---|---|---|
| δH, m (J Hz) | δC | δH, m (J Hz) | δC | δH, m (J Hz) | δC | δH, m (J Hz) | δC | ||
| 1 | 169.3 | 169.2 | 33 | 4.17, td (10.0, 2.3) | 70.2 | 4.17, td (10.0, 2.0) | 70.2 | ||
| 2 | 128.8 | 128.9 | 34 | 1.69 b | 43.2 | 1.70 b | 43.2 | ||
| 3 | 6.80, m | 144.4 | 6.79, m | 144.2 | 35 | 4.02, m | 66.5 | 4.01, m | 66.4 |
| 4 | 2.26 a, m | 27.6 | 2.25 a, m | 27.6 | 36 | 1.53 g | 46.4 | 1.52 f | 46.3 l |
| 5 | a 1.42, m | 34.5 | a 1.41, m | 34.4 i | 37 | 4.27, m | 69.7 | 4.27, m | 69.7 |
| b 1.57, m | b 1.56, m | 38 | 5.58, dd (15.4, 5.5) | 138.2 | 5.56, dd (15.5, 5.7) | 138.0 | |||
| 6 | 1.67 b, m | 36.0 | 1.65 b, m | 35.9 | 39 | 5.62, dd (15.4, 6.7) | 125.8 | 5.62, dd (15.0, 7.2) | 125.9 |
| 7 | 3.40, dd (9.6, 1.7) | 77.5 | 3.39, dd (9.6, 1.6) | 77.5 | 40 | a 2.30, dd (13.4, 6.7) | 34.5 | a 2.29, dd (13.5, 7.2) | 34.4 i |
| 8 | 1.75, m | 42.8 | 1.74, m | 42.8 | b 2.45, dd (13.4, 5.3) | b 2.44, dd (13.5, 5.1) | |||
| 9 | 3.80 c | 74.7 k | 3.80 c | 74.7 j | 41 | 5.10, m | 75.6 l | 5.11, m | 75.7 k |
| 10 | a 1.40 d | a 1.38 | 33.5 | 42 | 1.99, q (6.9) | 43.7 | 1.97, q (6.8) | 43.5 | |
| b 1.60 e | b 1.59 d | 43 | 3.52, ddd (9.8, 5.6, 1.9) | 72.6 | 3.50, ddd (8.4, 5.5, 1.8) | 72.7 | |||
| 11 | a 2.07, m | 30.40 | a 2.07, m | 30.4 | 44 | a 1.40 d | 30.44 | a 1.31 | 31.0 |
| b 2.24 a | b 2.24 a | b 1.63 e | b 1.54 f | ||||||
| 12 | 5.49 f | 131.6 | 5.49 e | 131.6 | 45 | a 1.67 b | 25.6 | a 1.48 f | 27.1 |
| 13 | 5.45 f | 134.0 | 5.46 e | 134.0 | b 1.82, m | b 1.68 b | |||
| 14 | 2.19, q (6.7) | 44.1 | 2.19, q (6.6) | 44.0 | 46 | 2.93, m | 40.8 | 3.15, m | 40.4 |
| 15 | 3.87, dd (6.7, 5.5) | 76.7 | 3.87, dd (6.6, 5.3) | 76.7 | 46-NHCOCH3 | --- | --- | --- | 173.2 |
| 16 | 5.49 f | 132.1 | 5.46 e | 132.1 | 46-NHCOCH3 | --- | --- | 1.92, s | 22.6 |
| 17 | 5.50 f | 134.7 | 5.49 e | 134.6 | 47 | 1.85, s | 12.7 | 1.85, s | 12.7 |
| 18 | 2.34, m | 41.2 | 2.33, m | 41.2 | 48 | 0.88, d (6.8) | 12.6 | 0.88, d (6.8) | 12.5 |
| 19 | 3.72, d (8.5) | 83.4 | 3.71, d | 83.4 | 49 | 0.78, d (6.9) | 12.0 | 0.78, d (7.5) | 12.0 |
| 20 | --- | 145.5 | --- | 145.6 | 50 | 0.97, d (6.8) | 16.1 | 0.98, d (6.9) | 16.1 |
| 21 | 5.30, d (9.6) | 124.1 | 5.30, d (9.3) | 124.0 | 51 | 1.11, d (6.6) | 17.6 | 1.11, d (6.6) | 17.6 |
| 22 | 4.39, dd (9.6, 3.5) | 75.6 l | 4.39, dd (9.6, 3.6) | 75.7 k | 52 | 1.71, s | 12.2 | 1.71, s | 12.2 |
| 23 | 3.96, dd (8.3, 3.5) | 71.7 | 3.96, dd (8.0, 3.4) | 71.7 | 53 | 0.94 h, d (6.9) | 10.1 | 0.94, d (6.9) | 10.1 |
| 24 | 1.49, m | 41.5 | 1.49, m | 41.46 | 54 | 0.79, d (7.0) | 11.5 | 0.79, d (7.5) | 11.5 |
| 25 | 4.08, m | 66.0 | 4.08, m | 66.0 | 55 | 0.94 h, d (6.9) | 10.6 | 0.92, d (6.9) | 10.8 |
| 26 | a 1.53 g | 46.4 | a 1.53 f | 46.3 l | 1′ | 4.83, d (1.5) | 97.8 | 4.83, d (1.3) | 97.8 |
| b 1.62 e | b 1.62 d | 2′ | 3.76 i | 72.4 * | 3.77 g | 72.4 * | |||
| 27 | 4.06, m | 69.1 | 4.07, m | 69.1 | 3′ | 3.76 i | 72.5 * | 3.75 g | 72.6 * |
| 28 | 1.44, m | 42.5 | 1.44, m | 42.55 | 4′ | 3.63 j | 68.7 | 3.63 h | 68.7 |
| 29 | 3.80 c | 75.1 | 3.80 c | 75.1 | 5′ | 3.63 j | 74.7 k | 3.63 h | 74.7 j |
| 30 | 1.63 e | 40.8 | 6′ | a 3.72, dd (11.5, 3.6) | 62.9 | a 3.72 | 62.9 | ||
| 31 | 3.98, dd (9.5, 1.7) | 73.5 | 3.98, dd (9.9, 1.7) | 73.5 | b 3.84, dd (11.5, 1.3) | b 3.84 | |||
| 32 | 1.69 b | 41.75 | 1.68 b | 41.7 | |||||
| Strain | Desertomycin A (1) | Desertomycin H (2) |
|---|---|---|
| Escherichia coli | >128 | >128 |
| Enterococcus faecalis | 64 | >128 |
| Mycobacterium luteus | 16 | >128 |
| Staphylococcus aureus (MSSA) 1 | 32 | >128 |
| Staphylococcus aureus (VISA5) 2 | 64 | >128 |
| Strain | Genotype | Source/Reference |
|---|---|---|
| PP655 | Bacillus subtilis 168 dal-1 sigB::erm (from AG232 [53]) | Alan Grossman |
| PP662 | Escherichia coli F-, Δ(araA-leu)7697, [araD139]B/r, Δ(codB-lacI)3, galK16, galE15(GalS), λ-, e14-, dadX100::FRT, relA1, rpsL150(strR), spoT1, alr-100::FRT, mcrB1 | E. coli Stock Center [54] |
| PP663 | Cornybacterium glutamicum ATCC 13869 | David Sherman |
| PP664 | Rhodococcus erythropolis B-16025. | David Sherman |
| PP665 | Klebsiella pneumoniae ATCC 13883 | ATCC |
| PP666 | Pseudomonas aeruginosa ATCC 27853 | ATCC |
| PP667 | Staphylococcus aureus ATCC 25923 | ATCC |
| PP673 | Mycobacterium smegmatis MC2155 | Miriam Braunstein |
| PP740 | Escherichia coli DH5α pGDP3_aph(3”)-Ia | AddGene |
| PP741 | Escherichia coli DH5α pGDP3_rmtB | AddGene |
| PP742 | Escherichia coli DH5α pGDP3_apmA | AddGene |
| PP743 | Escherichia coli DH5α pGDP3_aph(9)-Ia | AddGene |
| PP744 | Escherichia coli DH5α pGDP3_NDM-1 | AddGene |
| PP745 | Escherichia coli DH5α pGDP3_ermC | AddGene |
| PP746 | Escherichia coli DH5α pGDP3_vatD | AddGene |
| PP747 | Escherichia coli DH5α pGDP3_stat | AddGene |
| PP748 | Escherichia coli DH5α pGDP3_tet(A) | AddGene |
| PP749 | Escherichia coli DH5α pGDP3_cat | AddGene |
| PP750 | Escherichia coli DH5α pGDP3_fosA | AddGene |
| PP751 | Escherichia coli DH5α pGDP3_arr | AddGene |
| PP752 | Escherichia coli DH5α pGDP3_uvrA | AddGene |
| PP753 | Escherichia coli DH5α pGDP3_vph | AddGene |
| PP754 | Escherichia coli DH5α pGDP3_MCR-1 | AddGene |
| PP771 | Enterococcus faecalis ATCC29212 | ATCC |
| PP788 | Micrococcus luteus | This study |
| PAP57 | Streptomyces sp. PAP57 | This study |
| PAP58 | Streptomyces sp. PAP58 | This study |
| PAP60 | Streptomyces sp. PAP60 | This study |
| PAP61 | Streptomyces sp. PAP61 | This study |
| PAP62 | Streptomyces sp. PAP62 | This study |
| PAP117 | Streptomyces sp. PAP117 | This study |
| PAP124 | Streptomyces sp. PAP124 | This study |
| PAP133 | Streptomyces sp. PAP133 | This study |
| PAP143 | Streptomyces sp. PAP143 | This study |
| PAP163 | Streptomyces sp. PAP163 | This study |
| PAP165 | Pseudomonas sp. PAP165 | This study |
| PAP181 | Streptomyces sp. PAP181 | This study |
| PAP203 | Paenibacillus sp. PAP203 | This study |
| MMR14 | Streptomyces sp. MMR14 | This study |
| TE-Ec | Escherichia coli ATCC 1775 | Tiny EarthTM |
| TE-Bs | Bacillus subtilis | Tiny EarthTM |
| VISA5 | Vancomycin-intermediate resistant Staphylococcus aureus | AR Bank |
| CRE128 | Carbapenem-resistant Escherichia coli | AR Bank |
| CRE143 | Carbapenem-resistant Klebsiella pneumoniae | AR Bank |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohamed, O.G.; Dorandish, S.; Lindow, R.; Steltz, M.; Shoukat, I.; Shoukat, M.; Chehade, H.; Baghdadi, S.; McAlister-Raeburn, M.; Kamal, A.; et al. Identification of a New Antimicrobial, Desertomycin H, Utilizing a Modified Crowded Plate Technique. Mar. Drugs 2021, 19, 424. https://doi.org/10.3390/md19080424
Mohamed OG, Dorandish S, Lindow R, Steltz M, Shoukat I, Shoukat M, Chehade H, Baghdadi S, McAlister-Raeburn M, Kamal A, et al. Identification of a New Antimicrobial, Desertomycin H, Utilizing a Modified Crowded Plate Technique. Marine Drugs. 2021; 19(8):424. https://doi.org/10.3390/md19080424
Chicago/Turabian StyleMohamed, Osama G., Sadaf Dorandish, Rebecca Lindow, Megan Steltz, Ifrah Shoukat, Maira Shoukat, Hussein Chehade, Sara Baghdadi, Madelaine McAlister-Raeburn, Asad Kamal, and et al. 2021. "Identification of a New Antimicrobial, Desertomycin H, Utilizing a Modified Crowded Plate Technique" Marine Drugs 19, no. 8: 424. https://doi.org/10.3390/md19080424
APA StyleMohamed, O. G., Dorandish, S., Lindow, R., Steltz, M., Shoukat, I., Shoukat, M., Chehade, H., Baghdadi, S., McAlister-Raeburn, M., Kamal, A., Abebe, D., Ali, K., Ivy, C., Antonova, M., Schultz, P., Angell, M., Clemans, D., Friebe, T., Sherman, D., ... Tripathi, A. (2021). Identification of a New Antimicrobial, Desertomycin H, Utilizing a Modified Crowded Plate Technique. Marine Drugs, 19(8), 424. https://doi.org/10.3390/md19080424