Epigenetic Manipulation Induced Production of Immunosuppressive Chromones and Cytochalasins from the Mangrove Endophytic Fungus Phomopsis asparagi DHS-48
Abstract
:1. Introduction
2. Results
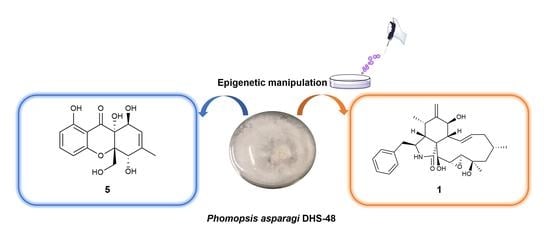
2.1. Epigenetic Manipulation
2.2. Structure Elucidation of the New Compounds
2.3. Biological Activity of Compounds
3. Materials and Methods
3.1. General Procedures
3.2. Fungal Material
3.3. Epigenetic Manipulation and Culture Condition
3.4. Extraction Isolation
3.5. Theory and Calculation Details
3.6. Cytotoxicity Assay
3.7. Isolation and Culture of Spleen Lymphocytes
3.8. Cell Activity and Cell Proliferation
3.9. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Xu, J. Bioactive natural products derived from mangrove-associated microbes. Rsc. Adv. 2015, 5, 841–892. [Google Scholar] [CrossRef]
- Xu, J. Biomolecules Produced by Mangrove-Associated Microbes. Curr. Med. Chem. 2011, 18, 5224–5266. [Google Scholar] [CrossRef] [PubMed]
- Bugni, T.S.; Ireland, C.M. Marine-Derived Fungi: A Chemically and Biologically Diverse Group of Microorganisms. Nat. Prod. Rep. 2004, 21, 143–163. [Google Scholar] [CrossRef]
- Xu, J.; Yi, M.; Ding, L.; He, S. A Review of Anti-Inflammatory Compounds from Marine Fungi, 2000–2018. Mar. Drugs 2019, 17, 636. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.J.; Xu, B.F.; Guo, Y.W. Unusual Secondary Metabolites from the Mangrove Ecosystems: Structures, Bioactivities, Chemical, and Bio-syntheses. Mar. Drugs. 2022, 20, 535. [Google Scholar] [CrossRef] [PubMed]
- Choque, E.; Klopp, C.; Valiere, S.; Raynal, J.; Mathieu, F. Whole-Genome Sequencing of Aspergillus tubingensis G131 and Overview of Its Secondary Metabolism Potential. BMC Genom. 2018, 19, 200. [Google Scholar] [CrossRef]
- Nützmann, H.W.; Reyes-Dominguez, Y.; Scherlach, K.; Schroeckh, V.; Horn, F.; Gacek, A.; Schümann, J.; Hertweck, C.; Strauss, J.; Brakhage, A.A. Bacteria-Induced Natural Product Formation in the Fungus Aspergillus nidulans Requires Saga/Ada-Mediated Histone Acetylation. Proc. Natl. Acad. Sci. USA 2011, 108, 14282–14287. [Google Scholar] [CrossRef]
- Pfannenstiel, B.T.; Keller, N.P. On Top of Biosynthetic Gene Clusters: How Epigenetic Machinery Influences Secondary Metabolism in Fungi. Biotechnol. Adv. 2019, 37, 107345. [Google Scholar] [CrossRef]
- Rutledge, P.J.; Challis, G.L. Discovery of Microbial Natural Products by Activation of Silent Biosynthetic Gene Clusters. Nat. Rev. Microbiol. 2015, 13, 509–523. [Google Scholar] [CrossRef]
- Skellam, E. Strategies for Engineering Natural Product Biosynthesis in Fungi. Trends Biotechnol. 2019, 37, 916. [Google Scholar] [CrossRef] [Green Version]
- Bode, H.B.; Bethe, B.; Höfs, R.; Zeeck, A. Big Effects from Small Changes: Possible Ways to Explore Nature’s Chemical Diversity. Chem. BioChem. 2002, 3, 619–627. [Google Scholar] [CrossRef]
- Liu, Y.Z.; Lu, C.H.; Shen, Y.M. Guanacastane-Type Diterpenoids from Coprinus plicatilis. Phytochem. Lett. 2014, 7, 161–164. [Google Scholar] [CrossRef]
- Wang, W.J.; Li, D.Y.; Li, Y.C.; Hua, H.M.; Ma, E.L.; Li, Z.L. Caryophyllene Sesquiterpenes from the Marine-Derived Fungus Ascotricha sp. ZJ-M-5 by the One Strain–Many Compounds Strategy. J. Nat. Prod. 2014, 77, 1367–1371. [Google Scholar] [CrossRef]
- Keller, N.P. Fungal Secondary Metabolism: Regulation, Function and Drug Discovery. Nat. Rev. Microbiol. 2019, 17, 167–180. [Google Scholar] [CrossRef] [PubMed]
- Lyu, H.N.; Liu, H.W.; Keller, N.P.; Yin, W.B. Harnessing Diverse Transcriptional Regulators for Natural Product Discovery in Fungi. Nat. Prod. Rep. 2020, 37, 6–16. [Google Scholar] [CrossRef]
- Reyes, F.; Bills, G.F.; Durán-Patrón, R. Strategies for the Discovery of Fungal Natural Products. Front. Microbiol. 2022, 13, 897756. [Google Scholar]
- Berger, S.L.; Kouzarides, T.; Shiekhattar, R.; Shilatifard, A. An Operational Definition of Epigenetics. Genes Dev. 2009, 23, 781–783. [Google Scholar]
- Wu, J.S.; Shi, X.H.; Zhang, Y.H.; Yu, J.Y.; Fu, X.M.; Li, X.; Chen, K.X.; Guo, Y.W.; Shao, C.L.; Wang, C.Y. Co-Cultivation With 5-Azacytidine Induced New Metabolites From the Zoanthid-Derived Fungus Cochliobolus lunatus. Front. Chem. 2019, 7, 763–770. [Google Scholar]
- Mafezoli, J.; Xu, Y.M.; Hilário, F.; Freidhof, B.; Espinosa Artiles, P.; dos Santos, L.C.; de Oliveira, M.C.F.; Gunatilaka, A.A.L. Modulation of Polyketide Biosynthetic Pathway of the Endophytic Fungus, Anteaglonium sp. FL0768, by Copper (II) and Anacardic Acid. Phytochem. Lett. 2018, 28, 157–163. [Google Scholar]
- Niu, S.W.; Liu, D.; Shao, Z.Z.; Proksch, P.; Lin, W.H. Eremophilane-Type Sesquiterpenoids in a Deep-Sea Fungus Eutypella sp. Activated by Chemical Epigenetic Manipulation. Tetrahedron 2018, 74, 7310–7325. [Google Scholar]
- Sharma, V.; Singamaneni, V.; Sharma, N.; Kumar, A.; Arora, D.; Kushwaha, M.; Bhushan, S.; Jaglan, S.; Gupta, P. Valproic Acid Induces Three Novel Cytotoxic Secondary Metabolites in Diaporthe sp., an Endophytic Fungus from Datura inoxia Mill. Bioorgan. Med. Chem. Lett. 2018, 28, 2217–2221. [Google Scholar] [CrossRef] [PubMed]
- Shi, T.; Shao, C.L.; Liu, Y.; Zhao, D.L.; Cao, F.; Fu, X.M.; Yu, J.Y.; Wu, J.S.; Zhang, Z.K.; Wang, C.Y. Terpenoids From the Coral-Derived Fungus Trichoderma harzianum (XS-20090075) Induced by Chemical Epigenetic Manipulation. Front. Microbiol. 2020, 11, 572–583. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.X.; Fang, H.; Yin, C.P.; Wei, C.L.; Hu, J.W.; Zhang, Y.L. Antimicrobial Metabolites Produced by Penicillium mallochii CCH01 Isolated From the Gut of Ectropis oblique, Cultivated in the Presence of a Histone Deacetylase Inhibitor. Front. Microbiol. 2019, 10, 2186–2193. [Google Scholar] [CrossRef] [PubMed]
- Asai, T.; Chung, Y.M.; Sakurai, H.; Ozeki, T.; Chang, F.R.; Wu, Y.C.; Yamashita, K.; Oshima, Y. Highly Oxidized Ergosterols and Isariotin Analogs from an Entomopathogenic Fungus, Gibellula formosana, Cultivated in the Presence of Epigenetic Modifying Agents. Tetrahedron 2012, 68, 5817–5823. [Google Scholar] [CrossRef]
- Niu, S.W.; Liu, D.; Shao, Z.Z.; Liu, J.R.; Fan, A.L.; Lin, W.H. Chemical Epigenetic Manipulation Triggers the Production of Sesquiterpenes from the Deep-Sea Derived Eutypella Fungus. Phytochemistry 2021, 192, 112978. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.S.; Yao, G.S.; Shi, X.H.; Rehman, S.U.; Xu, Y.; Fu, X.M.; Zhang, X.L.; Liu, Y.; Wang, C.Y. Epigenetic Agents Trigger the Production of Bioactive Nucleoside Derivatives and Bisabolane Sesquiterpenes From the Marine-Derived Fungus Aspergillus versicolor. Front. Microbiol. 2020, 11, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Beau, J.; Mahid, N.; Burda, W.N.; Harrington, L.; Shaw, L.N.; Mutka, T.; Kyle, D.E.; Barisic, B.; Van Olphen, A.; Baker, B.J. Epigenetic tailoring for the production of anti-infective cytosporones from the marine fungus Leucostoma persoonia. Mar. Drugs 2012, 10, 762–774. [Google Scholar] [CrossRef]
- Demers, D.H.; Knestrick, M.A.; Fleeman, R.; Tawfik, R.; Azhari, A.; Souza, A.; Baker, B.J. Exploitation of mangrove endophytic fungi for infectious disease drug discovery. Mar. Drugs 2018, 16, 376. [Google Scholar] [CrossRef]
- Farr, D.F.; Castlebury, L.A.; Rossman, A.Y. Morphological and molecular characterization of Phomopsis vaccinii and additional isolates of Phomopsis from blueberry and cranberry in the eastern United States. Mycologia 2002, 94, 494–504. [Google Scholar] [CrossRef]
- Yan, B.C.; Wang, W.G.; Hu, D.B.; Sun, X.; Kong, L.M.; Li, X.N.; Du, X.; Luo, S.H.; Liu, Y.; Li, Y.; et al. Phomopchalasins A and B, two cytochalasans with polycyclic-fused skeletons from the endophytic fungus Phomopsis sp. shj2. Org. Lett. 2016, 18, 1108–1111. [Google Scholar] [CrossRef]
- Shang, Z.; Raju, R.; Salim, A.A.; Khalil, Z.G.; Capon, R.J. Cytochalasins from an australian marine sediment-derived Phomopsis sp. (cmb-m0042f): Acid-mediated intramolecular cycloadditions enhance chemical diversity. J. Org. Chem. 2017, 82, 9704–9709. [Google Scholar] [CrossRef] [PubMed]
- Wagenaar, M.M.; Clardy, J. New Antibiotic and Cytotoxic Dimers Produced by the Fungus Phomopsis longicolla Isolated from an Endangered Mint. J. Nat. Prod. 2001, 64, 1006–1009. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.J.; Yang, H.Y.; Li, J.; Liu, X.; Ye, L.; Kong, W.S.; Tang, S.Y.; Du, G.; Liu, Z.H.; Zhou, M.; et al. Isopentylated diphenyl ether derivatives from the fermentation products of an endophytic fungus Phomopsis fukushii. J. Antibiot. 2018, 71, 359–362. [Google Scholar] [CrossRef] [PubMed]
- Elsässer, B.; Krohn, K.; Flörke, U.; Root, N.; Aust, H.J.; Draeger, S.; Schulz, B.; Antus, S.; Kurtán, T. X-ray Structure Determination, Absolute Configuration and Biological Activity of Phomoxanthone A. Eur. J. Org. Chem. 2005, 2005, 4563–4570. [Google Scholar] [CrossRef]
- Horn, W.S.; Simmonds, M.S.J.; Schwartz, R.E.; Blaney, W.M. Phomopsichalasin, a novel antimicrobial agent from an endophytic Phomopsis sp. Tetrahedron 1995, 51, 3969–3978. [Google Scholar] [CrossRef]
- Xie, S.; Wu, Y.; Qiao, Y.; Guo, Y.; Wang, J.; Hu, Z.; Zhang, Q.; Li, X.; Huang, J.; Zhou, Q.; et al. Protoilludane, illudalane, and botryane sesquiterpenoids from the endophytic fungus Phomopsis sp. tj507a. J. Nat. Prod. 2018, 81, 1311–1320. [Google Scholar] [CrossRef]
- Tan, Q.W.; Fang, P.H.; Ni, J.C.; Gao, F.; Chen, Q.J. Metabolites Produced by an Endophytic Phomopsis sp. and Their Anti-TMV Activity. Molecules 2017, 22, 2073. [Google Scholar] [CrossRef]
- Feng, Z.; Zhang, X.X.; Wu, J.W.; Wei, C.W.; Feng, T.; Zhou, D.D.; Wen, Z.C.; Xu, J. Immunosuppressive Cytochalasins from the Mangrove Endophytic Fungus Phomopsis asparagi DHS-48. Mar. Drugs 2022, 20, 526. [Google Scholar] [CrossRef]
- Xu, Z.Y.; Xiong, B.X.; Xu, J. Chemical investigation of secondary metabolites produced by mangrove endophytic fungus Phyllosticta capitalensis. Nat. Prod. Res. 2019, 35, 1561–1565. [Google Scholar] [CrossRef]
- Hemberger, Y.; Xu, J.; Wray, V.; Proksch, P.; Wu, J.; Bringmann, G. Pestalotiopens A and B: Stereochemically challenging flflexible sesquiterpene-cyclopaldic acid Hybrids from Pestalotiopsis sp. Chem. Eur. J. 2013, 19, 15556–15564. [Google Scholar] [CrossRef]
- Deng, Q.; Li, G.; Sun, M.Y.; Yang, X.; Xu, J. A new antimicrobial sesquiterpene isolated from endophytic fungus Cytospora sp. from the Chinese mangrove plant Ceriops tagal. Nat. Prod. Res. 2020, 34, 1404–1408. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.Y.; Zhou, D.D.; Wu, J.W.; Zhou, J.; Xu, J. Sdy-1 Executes Antitumor Activity in HepG2 and HeLa Cancer Cells by Inhibiting the Wnt/β-Catenin Signaling Pathway. Mar. Drugs 2022, 20, 125. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.W.; Deng, Q.; Sun, M.Y.; Xu, J. Cytospyrone and Cytospomarin: Two New Polyketides Isolated from Mangrove Endophytic Fungus, Cytospora sp. Molecules 2020, 25, 4224. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Kjer, J.; Sendker, J.; Wray, V.; Guan, H.; Edrada, R.; Lin, W.H.; Wu, J.; Proksch, P. Chromones from the Endophytic Fungus Pestalotiopsis sp. Isolated from the Chinese Mangrove Plant Rhizophora Mucronata. J. Nat. Prod. 2009, 72, 662–665. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Li, G.; Deng, Q.; Zheng, D.Y.; Yang, X.B.; Xu, J. Cytotoxic Constituents from the Mangrove Endophytic Pestalotiopsis sp. Induce G (0)/G (1) Cell Cycle Arrest and Apoptosis in Human Cancer Cells. Nat. Prod. Res. 2018, 32, 2968–2972. [Google Scholar] [CrossRef]
- Wei, C.W.; Sun, C.X.; Feng, Z.; Zhang, X.X.; Xu, J. Four New Chromones from the Endophytic Fungus Phomopsis asparagi DHS-48 Isolated from the Chinese Mangrove Plant Rhizophora mangle. Mar. Drugs 2021, 19, 348. [Google Scholar] [CrossRef]
- Xu, Z.Y.; Wu, X.; Li, G.; Feng, Z.; Xu, J. Pestalotiopisorin B, a New Isocoumarin Derivative from the Mangrove Endophytic Fungus Pestalotiopsis sp. HHL101. Nat. Prod. Res. 2020, 34, 1002–1007. [Google Scholar] [CrossRef]
- Zhang, X.X.; Li, G.; Deng, Q.; Xu, Z.Y.; Cen, J.R.; Xu, J. Vomifoliol Isolated from Mangrove Plant Ceriops Tagal Inhibits the NFAT Signaling Pathway with CN as the Target Enzyme in vitro. Bioorgan. Med. Chem. Lett. 2021, 48, 128235. [Google Scholar] [CrossRef]
- Chen, S.H.; Guo, H.; Jiang, M.H.; Wu, Q.L.; Li, J.; Shen, H.J.; Liu, L. Mono- and Dimeric Xanthones with Anti-Glioma and Anti-Inflammatory Activities from the Ascidian-Derived Fungus Diaporthe sp. SYSU-MS4722. Mar. Drugs 2022, 20, 51. [Google Scholar] [CrossRef]
- Huang, M.X.; Li, J.; Liu, L.; Yin, S.; Wang, J.; Lin, Y.C. Phomopsichin A-D; four new chromone derivatives from mangrove endophytic fungus Phomopsis sp. 33. Mar. Drugs 2016, 14, 215. [Google Scholar] [CrossRef]
- Wijeratne, E.K.; Bashyal, B.P.; Gunatilaka, M.K.; Arnold, A.E.; Gunatilaka, A.L. Maximizing chemical diversity of fungal metabolites: Biogenetically related heptaketides of the endolichenic fungus Corynespora sp. J. Nat. Prod. 2010, 73, 1156–1159. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Liu, Z.M.; Chen, Y.C.; Tan, H.B.; Li, S.N.; Li, H.H. Chromone-derived polyketides from the deep-sea fungus Diaporthe phaseolorum fs431. Mar. Drugs 2019, 17, 182. [Google Scholar] [CrossRef] [PubMed]
- Cole, R.J.; Wells, J.M.; Cox, R.H.; Cutler, H.G.J. Isolation and biological properties of deacetylcytochalasin H from Phomopsis sp. Agric. Food Chem. 1981, 29, 205–206. [Google Scholar] [CrossRef]
- Bisht, R.; Bhattacharyya, A.; Shrivastava, A.; Saxena, P. An overview of the medicinally important plant type III PKS derived polyketides. Front. Plant Sci. 2021, 12, 746908. [Google Scholar] [CrossRef]
- Rönsberg, D.; Debbab, A.; Mándi, A.; Vasylyeva, V.; Böhler, P.; Stork, B.; Proksch, P. Pro-apoptotic and immunostimulatory tetrahydroxanthone dimers from the endophytic fungus Phomopsis longicolla. J. Org. Chem. 2013, 78, 12409–12425. [Google Scholar] [CrossRef] [PubMed]
- Lösgen, S.; Schlörke, O.; Meindl, K.; Herbst-Irmer, R.; Zeeck, A. Structure and biosynthesis of chaetocyclinones, new polyketides produced by an endosymbiotic fungus. Eur. J. Org. Chem. 2010, 2007, 2191–2196. [Google Scholar] [CrossRef]
- Zhou, J.; Diao, X.; Wang, T.; Chen, G.; Lin, Q.; Yang, X.; Xu, J. Phylogenetic diversity and antioxidant activities of culturable fungal endophytes associated with the mangrove species Rhizophora stylosa and R. mucronata in the South China Sea. PLoS ONE 2018, 13, e0197359. [Google Scholar] [CrossRef] [Green Version]
- Liu, G.; Xu, Z.N.; Cen, P.L. A Morphologically Structured Model for Mycelial Growth and Secondary Metabolite Formation. Chin. J. Chem. Eng. 2000, 8, 46–51. [Google Scholar]











| Position | 1 | 2 | ||
|---|---|---|---|---|
| δC Type | δH (J in Hz) | δC Type | δH (J in Hz) | |
| 1 | 163.2, C | 163.4, C | ||
| 2 | 109.3, CH | 6.43, d, 8.2 | 109.7, CH | 6.44, d, 8.2 |
| 3 | 138.9, CH | 7.38, t, 8.2 | 139.0, CH | 7.39, t, 8.2 |
| 4 | 108.7, CH | 6.52, d, 8.2 | 109.5, CH | 6.55, d, 8.2 |
| 4a | 160.8, C | 160.3, C | ||
| 5 | 74.3, CH | 4.69, brs | 74.6, CH | 4.29, m |
| 6 | 140.5, C | 29.2, CH | 2.34, m | |
| 7 | 122.3, CH | 5.61, dq, 4.8,1.7 | 32.0, CH2 | 1.57, dt, 14.8, 2.8 |
| 2.43, m | ||||
| 8 | 67.9, CH | 4.55, d, 4.8 | 68.6, CH | 4.44, t, 3.6 |
| 8a | 74.5, C | 85.4, C | ||
| 9 | 197.2, C | 196.2, C | ||
| 9a | 108.4, C | 108.6, C | ||
| 10a | 86.5, C | 76.4, C | ||
| 11 | 19.2, CH3 | 1.86, d, 1.7 | 18.1, CH3 | 1.32, d, 7.8 |
| 12 | 64.1, CH2 | Ha 4.12, d, 13.2 | 60.5, CH2 | Ha 3.83, d, 13.5 |
| Hb 4.03, d, 13.2 | Hb 4.28, m | |||
| Position | 5 | 6 | ||
|---|---|---|---|---|
| δC Type | δH (J in Hz) | δC Type | δH (J in Hz) | |
| 1 | 178.4, C | 178.7, C | ||
| 3 | 55.3, CH | 3.37, dd, 6.1, 3.5 | 55.1, CH | 3.31, m |
| 4 | 51.9, CH | 2.46, dd, 5.1, 3.5 | 50.8, CH | 2.61, m |
| 5 | 33.8, CH | 2.75, m | 34.0, CH | 2.92, m |
| 6 | 151.7, C | 151.9, C | ||
| 7 | 72.7, CH | 3.82, d, 10.8 | 72.5, CH | 3.83, d, 10.7 |
| 8 | 45.8, CH | 2.89, d, 10.1 | 50.2, CH | 2.88, m |
| 9 | 56.1, C | 54.9, C | ||
| 10 | 44.8, CH2 | Ha 2.86, dd, 10.3, 6.2Hb 2.77, m | 46.6, CH2 | Ha 2.92, mHb 2.59, dd, 13.6, 8.2 |
| 11 | 13.8, CH3 | 0.80, d, 6.7 | 14, CH3 | 1.11, d, 6.7 |
| 12 | 113.1, CH2 | Ha 5.22, s | 113.1, CH2 | Ha 5.34, s |
| Hb 5.01, s | Hb 5.12, s | |||
| 13 | 131.2, CH | 5.72, dd, 15.5, 9.5 | 129.4, CH | 5.73, dd, 15.5, 9.8 |
| 14 | 135.4, CH | 5.54, m | 138.7, CH | 5.33, m |
| 15 | 43.7, CH2 | Ha 2.05, dd, 12.4, 5.6Hb 1.65, d, 12.4 | 45, CH2 | Ha 2.02, mHb 1.81, m |
| 16 | 28.7, CH | 1.87, m | 29.4, CH | 1.80, m |
| 17 | 48.7, CH2 | Ha 1.75, dd, 14.8, 3.2Hb 1.45, dd, 14.8, 3.8 | 55, CH2 | Ha 1.90, dd, 14.4, 3.4Hb 1.57, dd, 14.4, 3.3 |
| 18 | 73.6, C | 75.1, C | ||
| 19 | 63.1, CH | 3.21, d, 2.4 | 137.5, CH | 5.75, dd, 16.8, 2.2 |
| 20 | 57.7, CH | 2.99, m | 132.2, CH | 6.03, dd, 16.8, 2.6 |
| 22 | 26.2, CH3 | 1.02, d, 6.8 | 26.7, CH3 | 1.04, d, 6.6 |
| 23 | 23.5, CH3 | 0.92, s | 31.8, CH3 | 1.36, s |
| 1’ | 138.5, C | 137.5, C | ||
| 2’/6’ | 131.2, CH | 7.22, d, 7.2 | 131.1, CH | 7.15, dd, 8.0, 1.3 |
| 3’/5’ | 129.6, CH | 7.31, t, 7.2 | 129.5, CH | 7.32, dd, 8.0, 7.1 |
| 4’ | 128.0, CH | 7.25, dt, 7.2, 3.6 | 127.8, CH | 7.24, dd, 7.1, 1.3 |
| Compound | IC50 (µM) a | |
|---|---|---|
| ConA-Induced T-Cell Proliferation | LPS-Induced B-Cell Proliferation | |
| 1 | 42.35 ± 2.49 | 88.19 ± 2.59 |
| 8 | 14.71 ± 0.47 | 109.95 ± 5.68 |
| 2–7, 9–11 | - | - |
| cyclosporin A b | 4.39 ± 0.02 | 25.11 ± 0.43 |
| Compound | IC50 (µM) a | |
|---|---|---|
| HepG2 | Hela | |
| 5 | 59.14 ± 15.79 | 5.82 ± 0.82 |
| 1–4, 6–11 | - | - |
| Adriamycin b | - | 0.95 ± 0.61 |
| Fluorouracil c | 176 ± 28.8 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, T.; Wei, C.; Deng, X.; Chen, D.; Wen, Z.; Xu, J. Epigenetic Manipulation Induced Production of Immunosuppressive Chromones and Cytochalasins from the Mangrove Endophytic Fungus Phomopsis asparagi DHS-48. Mar. Drugs 2022, 20, 616. https://doi.org/10.3390/md20100616
Feng T, Wei C, Deng X, Chen D, Wen Z, Xu J. Epigenetic Manipulation Induced Production of Immunosuppressive Chromones and Cytochalasins from the Mangrove Endophytic Fungus Phomopsis asparagi DHS-48. Marine Drugs. 2022; 20(10):616. https://doi.org/10.3390/md20100616
Chicago/Turabian StyleFeng, Ting, Chengwen Wei, Xiaolin Deng, Dandan Chen, Zhenchang Wen, and Jing Xu. 2022. "Epigenetic Manipulation Induced Production of Immunosuppressive Chromones and Cytochalasins from the Mangrove Endophytic Fungus Phomopsis asparagi DHS-48" Marine Drugs 20, no. 10: 616. https://doi.org/10.3390/md20100616
APA StyleFeng, T., Wei, C., Deng, X., Chen, D., Wen, Z., & Xu, J. (2022). Epigenetic Manipulation Induced Production of Immunosuppressive Chromones and Cytochalasins from the Mangrove Endophytic Fungus Phomopsis asparagi DHS-48. Marine Drugs, 20(10), 616. https://doi.org/10.3390/md20100616