Computational Metabolomics Tools Reveal Subarmigerides, Unprecedented Linear Peptides from the Marine Sponge Holobiont Callyspongia subarmigera
Abstract
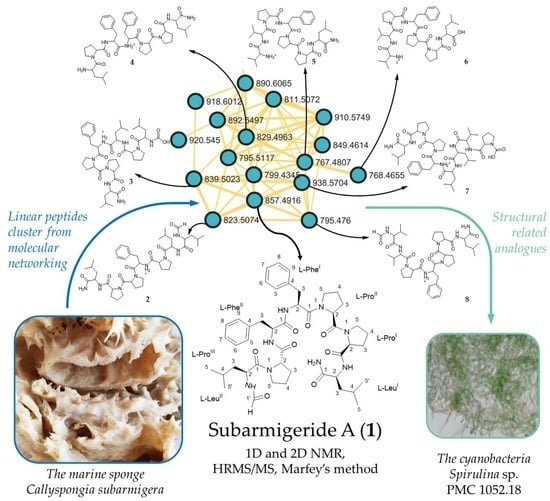
:1. Introduction
2. Results and Discussion
2.1. Structure Characterization of Subarmigeride A (1)
2.2. Putative Structure of Seven Additional Linear Peptides
2.3. Occurrence of the Linear Peptide Subarmigeride A (1) in the Previously Studied Cyanobacterium PMC 1052.18 from Guadeloupe
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Sponge Material
3.3. Sponge Extract Preparation
3.4. Isolation and Purification
3.5. NMR Data Acquisition and Processing
3.6. LC-MS2 Analyses of Extracts
3.7. Mass Spectrometry: LC-MS/MS Data Processing
3.8. Mass Spectrometry: Molecular Networking
3.9. Advanced Marfey’s Analysis
3.10. MASST Analysis
3.11. Mass Spectrometry: Comparison of LC-MS/MS Data of the Cyanobacterial Strain PMC 1052.18 and the Marine Sponge Callyspongia subarmigera
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Beniddir, M.A.; Le Moyec, L.; Triba, M.N.; Longeon, A.; Deville, A.; Blond, A.; Pham, V.C.; de Voogd, N.J.; Bourguet-Kondracki, M.-L. Metabolomics with multi-block modelling of mass spectrometry and nuclear magnetic resonance in order to discriminate Haplosclerida marine sponges. Anal. Bioanal. Chem. 2022, 414, 5929–5942. [Google Scholar] [CrossRef] [PubMed]
- Nothias, L.-F.; Petras, D.; Schmid, R.; Dührkop, K.; Rainer, J.; Sarvepalli, A.; Protsyuk, I.; Ernst, M.; Tsugawa, H.; Fleischauer, M.; et al. Feature-based molecular networking in the GNPS analysis environment. Nat. Methods 2020, 17, 905–908. [Google Scholar] [CrossRef] [PubMed]
- Olivon, F.; Allard, P.-M.; Koval, A.; Righi, D.; Genta-Jouve, G.; Neyts, J.; Apel, C.; Pannecouque, C.; Nothias, L.-F.; Cachet, X.; et al. Bioactive Natural Products Prioritization Using Massive Multi-Informational Molecular Networks. ACS Chem. Biol. 2017, 12, 2644–2651. [Google Scholar] [CrossRef] [PubMed]
- Olivon, F.; Apel, C.; Retailleau, P.; Allard, P.M.; Wolfender, J.L.; Touboul, D.; Roussi, F.; Litaudon, M.; Desrat, S. Searching for original natural products by molecular networking: Detection, isolation and total synthesis of chloroaustralasines. Org. Chem. Front. 2018, 5, 2171–2178. [Google Scholar] [CrossRef]
- Wolfender, J.-L.; Litaudon, M.; Touboul, D.; Queiroz, E.F. Innovative omics-based approaches for prioritisation and targeted isolation of natural products—new strategies for drug discovery. Nat. Prod. Rep. 2019, 36, 855–868. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fox Ramos, A.E.; Evanno, L.; Poupon, E.; Champy, P.; Beniddir, M.A. Natural products targeting strategies involving molecular networking: Different manners, one goal. Nat. Prod. Rep. 2019, 36, 960–980. [Google Scholar] [CrossRef] [PubMed]
- De Sousa, L.H.N.; de Araújo, R.D.; Sousa-Fontoura, D.; Menezes, F.G.; Araújo, R.M. Metabolities from Marine Sponges of the Genus Callyspongia: Occurrence, Biological Activity, and NMR Data. Mar. Drugs 2021, 19, 663. [Google Scholar] [CrossRef]
- Wang, M.; Jarmusch, A.K.; Vargas, F.; Aksenov, A.A.; Gauglitz, J.M.; Weldon, K.; Petras, D.; da Silva, R.; Quinn, R.; Melnik, A.V.; et al. Mass spectrometry searches using MASST. Nat. Biotechnol. 2020, 38, 23–26. [Google Scholar] [CrossRef] [PubMed]
- Duperron, S.; Beniddir, M.A.; Durand, S.; Longeon, A.; Duval, C.; Gros, O.; Bernard, C.; Bourguet-Kondracki, M.-L. New benthic cyanobacteria from guadeloupe mangroves as producers of antimicrobials. Mar. Drugs 2019, 18, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scarpato, S.; Teta, R.; Della Sala, G.; Pawlik, J.R.; Costantino, V.; Mangoni, A. New Tricks with an Old Sponge: Feature-Based Molecular Networking Led to Fast Identification of New Stylissamide L from Stylissa caribica. Mar. Drugs 2020, 18, 443. [Google Scholar] [CrossRef] [PubMed]
- Teta, R.; Marteinsson, V.T.; Longeon, A.; Klonowski, A.M.; Groben, R.; Bourguet-Kondracki, M.-L.; Costantino, V.; Mangoni, A. Thermoactinoamide A, an Antibiotic Lipophilic Cyclopeptide from the Icelandic Thermophilic Bacterium Thermoactinomyces vulgaris. J. Nat. Prod. 2017, 80, 2530–2535. [Google Scholar] [CrossRef]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [Green Version]
- Gantt, S.E.; McMurray, S.E.; Stubler, A.D.; Finelli, C.M.; Pawlik, J.R.; Erwin, P.M. Testing the relationship between microbiome composition and flux of carbon and nutrients in Caribbean coral reef sponges. Microbiome 2019, 7, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chambers, M.C.; Maclean, B.; Burke, R.; Amodei, D.; Ruderman, D.L.; Neumann, S.; Gatto, L.; Fischer, B.; Pratt, B.; Egertson, J.; et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012, 30, 918–920. [Google Scholar] [CrossRef] [PubMed]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Myers, O.D.; Sumner, S.J.; Li, S.; Barnes, S.; Du, X. One step forward for reducing false positive and false negative compound identifications from mass spectrometry metabolomics data: New algorithms for constructing extracted ion chromatograms and detecting chromatographic peaks. Anal. Chem. 2017, 89, 8696–8703. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Mi, Y.; Zhang, J.; He, S.; Yan, X. New Peptides Isolated from Marine Cyanobacteria, an Overview over the Past Decade. Mar. Drugs 2017, 15, 132. [Google Scholar] [CrossRef] [PubMed]


| Unit | Pos. | δC, Mult. | δH (J in Hz) | HMBC | NOESY a | |
|---|---|---|---|---|---|---|
| l-LeuI | NH | - | 7.71, d (8.22) | LeuI-1, ProI-CO | ProI-1, ProI-2-a, ProI-2-b, ProI-3-a, ProI-3-b, ProI-4-a | |
| 1 | 174.1, C | - | - | - | ||
| 2 | 50.7, CH | 4.16, ddd (6.72, 8.22, 14.93) | LeuI-CO, LeuI-2, LeuI-3, ProI-CO | ProI-1 | ||
| 3 | 40.7, CH2 | 1,47, m | LeuI-1, LeuI-3, LeuI-4, LeuI-4′ | - | ||
| 4 | 24.2, CH | 1.61, m | LeuI-2, LeuI-4, LeuI-4′ | ProI-2-a, ProI-2-b | ||
| 5 | 23.1, CH3 | 0.87, d (6.59) | LeuI-2, LeuI- 3, LeuI-4′ | - | ||
| 5′ | 21.5, CH3 | 0.83, d (6.59) | LeuI-2, LeuI- 3, LeuI-4 | - | ||
| NH2 | - | a. b. | 6.97, brs 7.17 brs | LeuI-CO, LeuI-1 | - - | |
| l-ProI | 1 | 171.2, C | - | - | - | |
| 2 | 59.7, CH | 4.31 dd (4.42, 8.28) | ProI-3 | LeuI-NH | ||
| 3 | 28.7, CH2 | a. b. | 1.80 m 2.02 m | ProI-CO | LeuI-NH, LeuI-3 LeuI-NH, LeuI-4 | |
| 4 | 24.6, CH2 | a. b. | 1.86 m 1.88 m | - - | LeuI-NH LeuI-NH | |
| 5 | 46.7, CH2 | a. b. | 3.52 m 3.67 m | ProI-2 | LeuI-NH, ProII-1, ProII-2-b ProII-1, ProII-2-b | |
| l-ProII | 1 | 170.1, C * | - | - | - | |
| 2 | 57.8, CH | 4.53, dd (4.45, 8.29) | - | PheI-1, ProI-3-a, ProI-3-b | ||
| 3 | 28.0, CH2 | a. b. | 1.84 m 2.09 m | - - | - ProI-4-a, ProI-4-b | |
| 4 | 24.3, CH2 | a. b. | 1.81 m 1.92 m | - - | - PheI-1 | |
| 5 | 46.6, CH2 | a. b. | 3.37 m 3.57 m | - - | PheI-1, PheI-2-b PheI-1 | |
| l-PheI | NH | - | 8.17, d (8.20) | PheII-CO | PheII-1, PheII-NH, LeuII-NH | |
| 1 | 168.9, C | - | - | - | ||
| 2 | 51.8, CH | 4.64, ddd (5.05, 8.20, 13.26) | PheI-CO | ProII-1, ProII-3-b, PheII-1, ProII-4-a, ProII-4-b | ||
| 3 | 36.7, CH2 | a. b | 2.76 dd (1.90, 8.20) 2.95 dd (4.87, 14.08) | PheI-CO, PheI-1, PheI-1, PheI-2/6, PheII-CO | - ProII-4-a | |
| 4 | 137.4, C | - | - | - | ||
| 5/9 | 129.2, CH | 7.25 b | PheI-2, PheI-4, PheI-6 | - | ||
| 6/8 | 128.1, CH | 7.23 b | PheI-1, PheI-5 | - | ||
| 7 | 126.3, CH | 7.17 b | PheI-2/6 | - | ||
| l-PheII | NH | - | 7.74, d (8.05) | ProIII-CO | PheI-1, ProIII-1, ProIII-2-a, ProIII-2-b, ProIII-3-b | |
| 1 | 170.3, C | - | - | - | ||
| 2 | 53.5, CH | 4.41, ddd (5.37, 8.05, 13.42) | PheII-CO, PheII-5-2 | PheI-1, PheI-NH, ProIII-1, LeuII-NH | ||
| 3 | 37.2, CH2 | a. b. | 2.78 dd (2.17, 10.02), 2.92 dd (5.42, 14.34) | PheII-CO, PheII-1, PheII-1, PheI-2/6 | - - | |
| 4 | 137.4, C | - | - | - | ||
| 5/9 | 129.2, CH | 7.15 b | PheII-2, PheII-4, PheII-6 | - | ||
| 6/8 | 127.9, CH | 7.20 b | PheII-1, PheII-5 | - | ||
| 7 | 126.1, CH | 7.16 b | PheII-2/6 | - | ||
| l-ProIII | 1 | 170.9, C | - | - | - | |
| 2 | 59.2, CH | 4.30, dd (4.45, 8.29) | - | PheII-NH, PheII-1, LeuII-1 | ||
| 3 | 28.7, CH2 | a. b. | 1.73 m 1.91 m | - - | PheII-NH PheII-NH | |
| 4 | 24.2, CH2 | a. b | 1.76 m 1.81 m | - - | - PheII-NH | |
| 5 | 46.6, CH2 | a. b. | 3.42 m 3.60 m | - - | LeuII-1, LeuII-2-a, LeuII-2-b LeuII-1, LeuII-7-2-a, LeuII-2-b | |
| l-LeuII | NH | - | 8.29 dd (1.31, 8.34) | LeuII-HCO | PheII-1, PheI-NH | |
| 1 | 169.8, C * | - | - | - | ||
| 2 | 47.1, CH | 4.58 ddd (4.31, 8.34, 12.09) | ProIII-CO | ProIII-1, LeuII-3-a, ProIII-3-b | ||
| 3 | 40.2, CH2 | a. b | 1.39 m 1.38 m | ProIII-3, ProIII-4′, ProIII-4′ | ProIII-4-a, ProIII-4-b ProIII-4-a, ProIII-4-b | |
| 4 | 24.0, CH | 1.57, m | - | - | ||
| 5 | 21.3, CH3 | 0.85, d (1.68) | ProIII-2, ProIII-3, ProIII-4′ | - | ||
| 5′ | 23.1, CH3 | 0.84, d (1.89) | ProIII-2, ProIII-3, ProIII-4 | - | ||
| HCO | 160.7, CH | 7.96 brs | LeuII-1 | - |
| Product Ion Assignment | (m/z) | Error, pm | Molecular Formula |
|---|---|---|---|
| CHO-Leu-Pro-Phe-Phe-Pro-Pro-Leu-NH2 + H+ | 857.4914 | 0.7 | C46H65N8O8 |
| Pro-Phe-Phe-Pro-Pro-Leu-NH2 + H+ | 716.4146 | −2.2 | C39H54N7O6 |
| Pro-Phe-Phe-Pro-Pro-Leu + H+ | 699.3862 | 0.4 | C39H51N6O6 |
| Pro-Phe-Phe-Pro-Pro-Leu + H+ | 671.3931 | −2.3 | C38H51N6O5 |
| Phe-Phe-Pro-Pro-Leu + H+ | 630.3298 | −1.9 | C35H44N5O6 |
| Phe-Phe-Pro-Pro + H+ | 533.2763 | −0.8 | C30H37N4O5 |
| CHO-Leu-Pro-Phe-Phe + H+ | 505.2813 | −0.8 | C29H37N4O4 |
| Phe-Phe-Pro-Pro-NH2 + H+ | 489.2506 | −1.9 | C28H33N4O4 |
| Phe-Pro-Pro-Leu-NH2 + H+ | 472.2919 | 0 | C25H38N5O4 |
| Phe-Pro-Pro-Leu-NH2 + H+ | 455.2659 | −1.4 | C25H35N4O4 |
| Phe-Phe-Pro + H+ | 392.1971 | −0.6 | C23H26N3O3 |
| CHO-Leu-Pro-Phe + H+ | 386.2075 | −0.2 | C21H28N3O4 |
| Pro fragment-Phe-Phe + H+ | 375.1700 | 0.9 | C23H23N2O3 |
| CHO-Leu-Pro-Phe + H+ | 358.2129 | −1 | C20H28N3O3 |
| Phe-Pro-Pro + H+ | 342.1805 | 2.2 | C19H24N3O3 |
| Pro-Pro-Leu-NH2 + H+ | 325.2232 | 0.6 | C16H29N4O3 |
| Pro-Pro-Leu + H+ | 308.1966 | 0.8 | C16H26N3O3 |
| Phe-Pro-NH2 + H+ | 245.1282 | 1.2 | C14H17N2O2 |
| Pro-Leu + H+ | 239.1387 | 1.4 | C12H19N2O3 |
| Pro-Leu-NH2 + H+ | 228.1702 | 1.8 | C11H22N3O2 |
| Pro-Leu + H+ | 217.1332 | 1.4 | C13H17N2O |
| Pro-Leu + H+ | 211.1437 | 1.7 | C11H19N2O2 |
| Pro-Pro + H+ | 195.1124 | 1.8 | C10H15N2O2 |
| Pro-Leu + H+ | 183.1485 | 3.5 | C10H19N2O |
| Pro-Pro + H+ | 167.1160 | 11.2 | C9H15N2O |
| Phe immonium fragment + H+ | 120.080173 | 5 | C8H10N |
| Leu immonium fragment + H+ | 86.096342 | 1 | C5H12N |
| Pro immonium fragment + H+ | 70.064503 | 8.9 | C4H8N |
| Subarmigeride at m/z * | Molecular Formula [M + H]+ | aa-1 | aa-2 | aa-3 | aa-4 | aa-5 | aa-6 | aa-7 | aa-8 |
|---|---|---|---|---|---|---|---|---|---|
| B (2) at 823.5078 | C43H67N8O8+ | NH2-Leu/Ile | Pro | Pro | Phe | Pro | Leu/ Ile | Leu/Ile-CHO | |
| C (3) at 839.5024 | C43H67N8O9+ | NH2-Leu/Ile | Pro | Pro | Phe | Leu/ Ile | Pro | Leu/Ile-COOH | |
| D (4) at 829.4962 | C45H65N8O7+ | NH2-Leu/Ile | Pro | Pro | Phe | Phe | Pro | Leu/Ile | |
| E (5) at 767.4807 | C40H63N8O7+ | NH2-Leu/Ile | Pro | Pro | Phe | Pro | Val | Val | |
| F (6) at 768.4650 | C40H63N7O8+ | Leu/Ile | Pro | Pro | Phe | Pro | Val | Val | |
| G (7) at 938.5698 | C48H76N9O10+ | NH2-Leu/Ile | Pro | Pro | Phe | Leu/ Ile | Leu/ Ile | Val | Pro-COOH |
| H (8) at 795.4763 | C41H63N8O8+ | NH2-Leu/Ile | Pro | Pro | Phe | Pro | Val | Val-CHO |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Castaldi, A.; Teta, R.; Esposito, G.; Beniddir, M.A.; De Voogd, N.J.; Duperron, S.; Costantino, V.; Bourguet-Kondracki, M.-L. Computational Metabolomics Tools Reveal Subarmigerides, Unprecedented Linear Peptides from the Marine Sponge Holobiont Callyspongia subarmigera. Mar. Drugs 2022, 20, 673. https://doi.org/10.3390/md20110673
Castaldi A, Teta R, Esposito G, Beniddir MA, De Voogd NJ, Duperron S, Costantino V, Bourguet-Kondracki M-L. Computational Metabolomics Tools Reveal Subarmigerides, Unprecedented Linear Peptides from the Marine Sponge Holobiont Callyspongia subarmigera. Marine Drugs. 2022; 20(11):673. https://doi.org/10.3390/md20110673
Chicago/Turabian StyleCastaldi, Andrea, Roberta Teta, Germana Esposito, Mehdi A. Beniddir, Nicole J. De Voogd, Sébastien Duperron, Valeria Costantino, and Marie-Lise Bourguet-Kondracki. 2022. "Computational Metabolomics Tools Reveal Subarmigerides, Unprecedented Linear Peptides from the Marine Sponge Holobiont Callyspongia subarmigera" Marine Drugs 20, no. 11: 673. https://doi.org/10.3390/md20110673
APA StyleCastaldi, A., Teta, R., Esposito, G., Beniddir, M. A., De Voogd, N. J., Duperron, S., Costantino, V., & Bourguet-Kondracki, M. -L. (2022). Computational Metabolomics Tools Reveal Subarmigerides, Unprecedented Linear Peptides from the Marine Sponge Holobiont Callyspongia subarmigera. Marine Drugs, 20(11), 673. https://doi.org/10.3390/md20110673