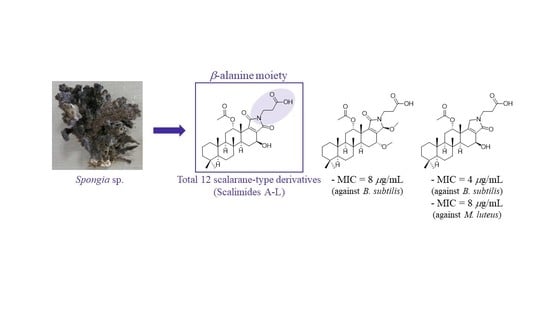
Isolation of Scalimides A–L: β-Alanine-Bearing Scalarane Analogs from the Marine Sponge Spongia sp.
Abstract
:1. Introduction
2. Results
2.1. Elucidation of the Structure
2.2. Biological Activities
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Biological Material
3.3. Extraction and Isolation
3.4. Assay
3.4.1. Antimicrobial Assays
3.4.2. Cytotoxicity Assays
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, K.; Gustafson, K.R. Sesterterpenoids: Chemistry, biology, and biosynthesis. Nat. Prod. Res. 2021, 38, 1251–1281. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.B.; Chen, H.Y.; Duan, S.; Zhu, Y.P.; Hu, B.; He, Y.; Cheng, S.T.; Jiao, B.H.; Liu, X.Y. Bioactive Scalarane—Type Sesterterpenoids from Marine Sources. Chem. Biodivers. 2022, 19, e202200049. [Google Scholar] [CrossRef] [PubMed]
- Máximo, P.; Lourenço, A. Marine sesterterpenes: An overview. Curr. Org. Chem. 2018, 22, 2381–2393. [Google Scholar] [CrossRef]
- Fattorusso, E.; Magno, S.; Santacroce, C.; Sica, D. Scalarin, a new pentacyclic C-25 terpenoid from the sponge Cacospongia scalaris. Tetrahedron 1972, 28, 5993–5997. [Google Scholar] [CrossRef]
- Abdelaleem, E.R.; Samy, M.N.; Desoukey, S.Y.; Liu, M.; Quinn, R.J.; Abdelmohsen, U.R. Marine natural products from sponges (Porifera) of the order Dictyoceratida (2013 to 2019); a promising source for drug discovery. RSC Adv. 2020, 10, 34959–34976. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, L.; Jung, J.H.; Zhang, S. Sesterterpenoids. Nat. Prod. Rep. 2007, 24, 1401–1429. [Google Scholar] [CrossRef]
- Gonzalez, M.A. Scalarane sesterterpenoids. Curr. Bioact. Compd. 2010, 6, 178–206. [Google Scholar] [CrossRef] [Green Version]
- Cafieri, F.; De Napoli, L.; Fattorusso, E.; Santacroce, C. Molliorin-B, a second scalarin-like pyrroloterpene from the sponge Cacospongia mollior. Experientia 1977, 33, 994–995. [Google Scholar] [CrossRef]
- Cafieri, F.; De Napoli, L.; Iengo, A.; Santacroce, C. Molliorin-c, a further pyrroloterpene present in the sponge Cacospongia mollior. Experientia 1978, 34, 300–301. [Google Scholar] [CrossRef]
- Cafieri, F.; De Napoli, L.; Iengo, A.; Santacroce, C. Minor pyrroloterpenoids from the marine sponge Cacospongia mollior. Experientia 1979, 35, 157–158. [Google Scholar] [CrossRef]
- Cafieri, F.; De Napoli, L.; Fattorusso, E.; Santacroce, C.; Sica, D. Molliorin-A: A unique scalarin-like pyrroloterpene from the sponge Cacospongia mollior. Tetrahedron Lett. 1977, 18, 477–480. [Google Scholar] [CrossRef]
- Hernández-Guerrero, C.J.; Zubia, E.; Ortega, M.J.; Carballo, J.L. Sesterterpene metabolites from the sponge Hyatella intestinalis. Tetrahedron 2006, 62, 5392–5400. [Google Scholar] [CrossRef]
- Jeon, J.-E.; Bae, J.; Lee, K.J.; Oh, K.-B.; Shin, J. Scalarane sesterterpenes from the sponge Hyatella sp. J. Nat. Prod. 2011, 74, 847–851. [Google Scholar] [CrossRef] [PubMed]
- Festa, C.; Cassiano, C.; D’Auria, M.V.; Debitus, C.; Monti, M.C.; De Marino, S. Scalarane sesterterpenes from Thorectidae sponges as inhibitors of TDP-43 nuclear factor. Org. Biomol. Chem. 2014, 12, 8646–8655. [Google Scholar] [CrossRef] [PubMed]
- Elhady, S.S.; El-Halawany, A.M.; Alahdal, A.M.; Hassanean, H.A.; Ahmed, S.A. A new bioactive metabolite isolated from the Red Sea marine sponge Hyrtios erectus. Molecules 2016, 21, 82. [Google Scholar] [CrossRef] [Green Version]
- Yang, I.; Lee, J.; Lee, J.; Hahn, D.; Chin, J.; Won, D.H.; Ko, J.; Choi, H.; Hong, A.; Nam, S.-J. Scalalactams A–D, scalarane sesterterpenes with a γ-lactam moiety from a Korean Spongia sp. Marine sponge. Molecules 2018, 23, 3187. [Google Scholar] [CrossRef] [Green Version]
- Kwon, O.-S.; Kim, D.; Kim, C.-K.; Sun, J.; Sim, C.J.; Oh, D.-C.; Lee, S.K.; Oh, K.-B.; Shin, J. Cytotoxic scalarane sesterterpenes from the sponge Hyrtios erectus. Mar. Drugs 2020, 18, 253. [Google Scholar] [CrossRef]
- Phan, C.-S.; Kamada, T.; Hamada, T.; Vairappan, C.S. Cytotoxic sesterterpenoids from bornean sponge Spongia sp. Rec. Nat. Prod. 2018, 12, 643–647. [Google Scholar] [CrossRef]
- Davis, R.; Capon, R. Two new scalarane sesterterpenes: Isoscalarafuran-A and-B, epimeric alcohols from a southern Australian Marine Sponge, Spongia hispida. Aust. J. Chem. 1993, 46, 1295–1299. [Google Scholar] [CrossRef]
- Nam, S.-J.; Ko, H.; Ju, M.K.; Hwang, H.; Chin, J.; Ham, J.; Lee, B.; Lee, J.; Won, D.H.; Choi, H. Scalarane sesterterpenes from a marine sponge of the genus Spongia and their FXR antagonistic activity. J. Nat. Prod. 2007, 70, 1691–1695. [Google Scholar] [CrossRef]
- Yang, I.; Nam, S.-J.; Kang, H. Two New Scalaranes from a Korean Marine Sponge Spongia sp. Nat. Prod. Sci. 2015, 21, 289–292. [Google Scholar] [CrossRef] [Green Version]
- Nam, S.-J.; Ko, H.; Shin, M.; Ham, J.; Chin, J.; Kim, Y.; Kim, H.; Shin, K.; Choi, H.; Kang, H. Farnesoid X-activated receptor antagonists from a marine sponge Spongia sp. Bioorg. Med. Chem. Lett. 2006, 16, 5398–5402. [Google Scholar] [CrossRef] [PubMed]






| Position | 1 | 2 | 3 | 4 | 5 | 6 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| δc | δH (J in Hz) | δc | δH (J in Hz) | δc | δH (J in Hz) | δc | δH (J in Hz) | δc | δH (J in Hz) | δc | δH (J in Hz) | |
| 1 | 40.9 | 1.59, m 0.63, m | 40.9 | 1.61, m 0.65, m | 40.9 | 1.59, m 0.63, m | 40.9 | 1.59, m 0.65, td (12.8, 3.7) | 40.9 | 1.60, m 0.65, m | 40.9 | 1.61, m 0.68, td (12.7, 3.8) |
| 2 | 19.5 | 1.65, m 1.41, m | 19.5 | 1.64, m 1.43, m | 19.5 | 1.63, m 1.40, m | 19.5 | 1.63, m 1.41, m | 19.5 | 1.65, m 1.42, m | 19.5 | 1.66, m 1.42, m |
| 3 | 43.2 | 1.38, m 1.15, m | 43.2 | 1.38, m 1.17, m | 43.2 | 1.38, m 1.15, td (13.1, 3.9) | 43.2 | 1.37, m 1.16, td (13.4, 4.0) | 43.2 | 1.38, m 1.17, td (13.1, 3.9) | 43.2 | 1.40, m 1.17, td (14.0, 13.5, 4.2) |
| 4 | 34.2 | 34.2 | 34.2 | 34.6 | 34.2 | 34.3 | ||||||
| 5 | 58.0 | 0.87, m | 58.0 | 0.92, m | 57.9 | 0.87, m | 58.0 | 0.88, m | 58.0 | 0.88, m | 58.0 | 0.91, m |
| 6 | 19.2 | 1.62, m 1.48, m | 19.2 | 1.63, m 1.48, m | 19.2 | 1.63, m 1.49, m | 19.2 | 1.63, m 1.49, m | 19.2 | 1.62, m 1.49, qd (13.2, 3.4) | 18.9 | 1.65, m 1.53, m |
| 7 | 42.4 | 1.88, m 1.03, m | 42.3 | 1.84, m 1.11, m | 42.3 | 1.89, dt (12.6, 3.5) 1.05, m | 42.4 | 1.88, dt (12.6, 3.0) 1.03, m | 42.4 | 1.88, dt (12.7, 3.5) 1.03, td (12.7, 3.9) | 41.9 | 2.03, m 1.03, m |
| 8 | 38.6 | 38.3 | 38.7 | 38.3 | 38.3 | 38.5 | ||||||
| 9 | 54.1 | 1.23, m | 54.3 | 1.32, m | 54.1 | 1.22, m | 54.3 | 1.25, dd (13.3, 2.3) | 54.3 | 1.26, dd (13.4, 2.3) | 53.4 | 1.31, m |
| 10 | 38.1 | 38.1 | 38.1 | 38.1 | 38.1 | 38.0 | ||||||
| 11 | 21.9 | 1.99, dd (12.6, 6.9) 1.78, ddd (15.2, 13.2, 2.4) | 21.9 | 2.04, m 1.82, m | 21.7 | 1.99, m 1.77, ddd (15.2, 13.2, 2.4) | 21.9 | 2.01, dt (15.1, 3.0) 1.78, ddd (15.2, 13.3, 2.5) | 21.9 | 2.01, dt (14.8, 2.8) 1.78, ddd (15.2, 13.2, 2.5) | 22.1 | 2.06, m 1.76, ddd (15.2, 13.2, 2.4) |
| 12 | 76.3 | 5.48, dd (3.5, 2.7) | 76.3 | 5.52, t (2.9) | 76.1 | 5.48, dd (3.2, 2.3) | 76.2 | 5.49, dd (3.5, 2.3) | 76.1 | 5.49, t (2.9) | 74.7 | 5.44, dd (3.5, 2.3) |
| 13 | 41.6 | 41.7 | 41.5 | 41.8 | 41.8 | 41.9 | ||||||
| 14 | 51.3 | 1.66, m | 46.6 | 2.03, m | 51.0 | 1.65, m | 46.9 | 1.92, m | 46.9 | 1.94, dd (12.9, 1.6) | 54.9 | 2.75, t (3.0) |
| 15 | 28.6 | 2.21, dd (12.8, 6.9) 1.57, m | 28.0 | 1.93, m 1.83, m | 25.5 | 2.32, dd (12.8, 7.1) 1.59, m | 23.5 | 2.11, dd (14.1, 1.6) 1.63, m | 23.5 | 2.11, m 1.62, m | 138.3 | 6.46, dd (9.7, 2.7) |
| 16 | 65.6 | 4.59, dd (9.4, 7.0) | 60.2 | 4.56, dd (4.3, 1.5) | 74.9 | 4.29, dd (9.1, 7.0) | 70.1 | 4.18, dd (4.0, 1.6) | 70.0 | 4.12, dd (4.0, 1.6) | 117.7 | 6.39, dd (9.7, 3.2) |
| 17 | 141.9 | 140.5 | 140.6 | 139.1 | 139.2 | 137.7 | ||||||
| 18 | 150.0 | 151.1 | 151.3 | 151.7 | 151.7 | 143.5 | ||||||
| 19 | 33.7 | 0.87, s | 33.8 | 0.87, s | 33.7 | 0.87, s | 33.7 | 0.87, s | 33.8 | 0.87, s | 33.8 | 0.88, s |
| 20 | 21.7 | 0.84, s | 21.7 | 0.85, s | 21.7 | 0.85, s | 21.8 | 0.85, s | 21.7 | 0.85, s | 21.8 | 0.85, s |
| 21 | 17.4 | 0.98, s | 17.6 | 0.96, s | 17.4 | 0.98, s | 17.6 | 0.97, s | 17.6 | 0.97, s | 19.3 | 1.09, s |
| 22 | 16.5 | 0.87, s | 16.4 | 0.88, s | 16.5 | 0.87, s | 16.4 | 0.88, s | 16.4 | 0.88, s | 16.4 | 0.89, s |
| 23 | 21.5 | 1.32, s | 20.1 | 1.21, s | 21.4 | 1.31, s | 19.9 | 1.22, s | 19.9 | 1.22, s | 17.0 | 1.11, s |
| 24 | 171.9 | 171.0 | 170.2 | 171.0 | 170.9 | 170.1 | ||||||
| 25 | 170.4 | 170.6 | 170.3 | 170.5 | 170.5 | 170.5 | ||||||
| 12-CH3CO | 172.1 | 172.1 | 171.8 | 172.0 | 171.9 | 171.9 | ||||||
| 12-CH3CO | 21.1 | 1.94, s | 21.1 | 1.94, s | 21.1 | 1.92, s | 21.1 | 1.94, s | 21.1 | 1.96, s | 21.2 | 1.99, s |
| 16-OCH3 | 58.0 | 3.52, s | 57.9 | 3.46, s | 57.9 | 3.46, s | ||||||
| 1′ | 34.5 | 3.69, m | 34.5 | 3.70, t (7.0) | 34.6 | 3.70, td (6.9, 1.7) | 34.6 | 3.70, td (6.9, 2.1) | 34.6 | 3.72, t (6.7) | 34.6 | 3.72, t (6.9) |
| 2′ | 33.5 | 2.56, td (6.9, 4.8) | 33.5 | 2.56, td (6.9, 1.5) | 33.4 | 2.56, td (6.9, 3.1) | 33.5 | 2.56, td (7.0, 5.0) | 33.8 | 2.56, td (6.7, 5.1) | 33.5 | 2.57, td (6.9, 3.1) |
| 3′ | 174.4 | 174.4 | 174.4 | 174.3 | 173.0 | 174.5 | ||||||
| 4′ | 52.3 | 3.63, s | ||||||||||
| Position | 7 | 8 | 9 | 10 | 11 | 12 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| δc | δH (J in Hz) | δc | δH (J in Hz) | δc | δH (J in Hz) | δc | δH (J in Hz) | δc | δH (J in Hz) | δc | δH (J in Hz) | |
| 1 | 40.9 | 1.60, m 0.64, m | 40.9 | 1.60, m 0.64, m | 40.9 | 1.61, m 0.65, m | 40.9 | 1.59, m 0.64, m | 40.9 | 1.60, m 0.68, m | 40.9 | 1.60, m 0.68, m |
| 2 | 19.5 | 1.64, m 1.40, m | 19.5 | 1.64, m 1.40, m | 19.5 | 1.66, m 1.42, m | 19.5 | 1.65, m 1.41, m | 19.6 | 1.66, m 1.42, m | 19.5 | 1.66, m 1.42, m |
| 3 | 43.2 | 1.38, m 1.16, m | 43.2 | 1.38, m 1.15, m | 43.2 | 1.40, m 1.18, m | 43.2 | 1.38, m 1.16, td (13.2, 4.0) | 43.2 | 1.39, dt (13.4, 3.8) 1.18, td (13.4, 4.1) | 43.2 | 1.39, m 1.18, td (13.2, 3.9) |
| 4 | 34.2 | 34.2 | 34.2 | 34.2 | 34.3 | 34.3 | ||||||
| 5 | 58.1 | 0.87, m | 58.0 | 0.88, m | 58.0 | 0.89, m | 58.0 | 0.88, m | 58.1 | 0.92, m | 58.1 | 0.91, m |
| 6 | 19.2 | 1.60, m 1.49, qd (13.0, 3.3) | 19.2 | 1.62, m 1.49, qd (13.0, 3.3) | 19.2 | 1.64, m 1.51, m | 19.2 | 1.63, m 1.49, m | 18.9 | 1.64, m 1.52, qd (12.8, 3.3) | 18.9 | 1.64, m 1.53, m |
| 7 | 42.4 | 1.88, m 1.03, td (12.8, 3.9) | 42.5 | 1.88, m 1.01, td (12.9, 4.1) | 42.6 | 1.90, m 1.03, dt (13.1, 7.0) | 42.5 | 1.89, m 1.04, m | 42.0 | 2.02, td (12.6, 3.4) 1.03, m | 41.9 | 2.02, m 1.03, m |
| 8 | 38.3 | 38.3 | 38.5 | 38.6 | 38.5 | 38.5 | ||||||
| 9 | 54.4 | 1.23, m | 54.4 | 1.23, m | 54.3 | 1.23, dd (13.3, 2.3) | 54.3 | 1.28, m | 53.5 | 1.35, dd (13.1, 2.4) | 53.5 | 1.34, m |
| 10 | 38.1 | 38.1 | 38.1 | 38.1 | 38.1 | 38.1 | ||||||
| 11 | 21.9 | 1.98, m 1.75 ddd (15.2, 13.2, 2.4) | 21.8 | 1.99, dt (14.8, 3.0) 1.75, ddd (15.2, 13.2, 2.5) | 21.8 | 1.99, m 1.76, ddd (15.2, 13.2, 2.5) | 22.3 | 1.91, m 1.79, m | 22.6 | 1.97, m 1.77, ddd (15.2, 13.0, 2.2) | 22.6 | 1.96, dd (14.8, 3.3) 1.77, m |
| 12 | 75.9 | 5.56, t (2.8) | 75.8 | 5.57, t (2.8) | 75.9 | 5.60, t (2.8) | 76.6 | 4.96, t (2.8) | 75.1 | 5.05, dd (3.7, 2.1) | 75.0 | 5.06, s |
| 13 | 40.7 | 41.1 | 40.9 | 42.3 | 43.4 | 43.4 | ||||||
| 14 | 47.5 | 1.79, m | 47.3 | 1.81, dd (13.0, 7.1) | 51.7 | 1.67, m | 50.9 | 1.68, m | 54.1 | 2.68, d (3.0) | 54.1 | 2.68, m |
| 15 | 22.8 | 2.12, d (14.4) 1.60, m | 22.5 | 2.13, d (14.5) 1.64, m | 28.9 | 2.14, dd (12.2, 6.6) 1.57, m | 28.6 | 2.15, dd (12.8, 7.0) 1.53, m | 130.3 | 6.03, dd (9.8, 2.6) | 130.3 | 6.03, dd (9.7, 2.6) |
| 16 | 71.3 | 4.02, dd (4.4, 1.3) | 71.7 | 3.91, dd (4.4, 1.4) | 68.6 | 4.44, dd (9.8, 6.6) | 66.4 | 4.49, dd (9.8, 6.7) | 118.8 | 6.25, dd (9.7, 3.3) | 118.8 | 6.24, dd (9.7, 3.3) |
| 17 | 151.4 | 148.8 | 153.9 | 133.4 | 130.3 | 130.3 | ||||||
| 18 | 142.8 | 145.6 | 140.1 | 162.3 | 159.8 | 159.8 | ||||||
| 19 | 33.8 | 0.88, s | 33.8 | 0.88, s | 33.7 | 0.88, s | 33.7 | 0.88, s | 33.8 | 0.88, s | 33.8 | 0.88, s |
| 20 | 21.8 | 0.85, s | 21.8 | 0.85, s | 21.7 | 0.86, s | 21.7 | 0.85, s | 21.8 | 0.86, s | 21.8 | 0.86, s |
| 21 | 17.5 | 0.96, s | 17.5 | 0.97, s | 17.6 | 0.98, s | 17.7 | 0.99, s | 19.2 | 1.10, s | 19.2 | 1.10, s |
| 22 | 16.5 | 0.88, s | 16.5 | 0.88, s | 16.4 | 0.88, s | 16.6 | 0.88, s | 16.4 | 0.89, s | 16.4 | 0.89, s |
| 23 | 19.9 | 1.16, s | 20.1 | 1.18, s | 21.6 | 1.26, s | 21.6 | 1.26, s | 17.5 | 1.06, s | 17.5 | 1.06, s |
| 24 | 81.7 | 5.39, d (1.9) | 87.0 | 5.44, m | 52.0 | 4.07, d (19.7) 3.97, d (19.8) | 172.1 | 171.6 | 173.9 | |||
| 25 | 170.0 | 170.3 | 171.7 | 50.4 | 4.06, d (19.6) 3.77, d (19.6) | 50.5 | 4.06, m | 50.4 | 4.09, m 4.01, m | |||
| 12-CH3CO | 172.2 | 172.1 | 172.3 | 172.5 | 172.0 | 172.4 | ||||||
| 12-CH3CO | 21.1 | 1.90, s | 21.1 | 1.90, s | 21.2 | 1.90, s | 21.2 | 2.01, s | 21.3 | 2.11, s | 21.3 | 2.13, s |
| 16-OCH3 | 57.4 | 3.43, s | 57.4 | 3.43, s | ||||||||
| 24-OCH3 | 50.5 | 3.04, s | ||||||||||
| 1′ | 36.2 | 3.68, m 3.53, dq (14.3, 7.2) | 36.6 | 3.69, m 3.40, m | 39.4 | 3.75, dt (13.4, 6.4) 3.58, dt (14.0, 6.7) | 39.3 | 3.74, m 3.58, dt (13.7, 6.4) | 39.5 | 3.73, dt (14.1, 6.3) 3.65, m | 39.4 | 3.74, m 3.66, m |
| 2′ | 34.2 | 2.63, dt (16.4, 7.3) 2.53, dt (16.4, 6.4) | 33.5 | 2.64, ddd (16.4, 8.0, 6.4) 2.52, dt (16.4, 6.1) | 34.1 | 2.60, dt (13.5, 6.6) | 34.0 | 2.59, m | 34.1 | 2.61, td (6.8, 6.3, 5.1) | 33.9 | 2.64, m |
| 3′ | 175.0 | 174.9 | 175.3 | 175.1 | 175.3 | 174.4 | ||||||
| 4′ | 52.3 | 3.65, s | ||||||||||
| Compounds | MIC (μg/mL) | MCF7 b | |||||
|---|---|---|---|---|---|---|---|
| Gram (+) Bacteria a | Gram (−) Bacteria a | ||||||
| A | B | C | D | E | F | EC50 (μM) | |
| 1 | 32 | 64 | 16 | >128 | >128 | >128 | 25.6 |
| 2 | >128 | >128 | 128 | >128 | >128 | >128 | 73.9 |
| 3 | 32 | 64 | 8 | >128 | >128 | >128 | 13.7 |
| 4 | 32 | 128 | 4 | >128 | >128 | >128 | 16.0 |
| 5 | >128 | >128 | >128 | >128 | >128 | >128 | 26.5 |
| 6 | 64 | 128 | 32 | >128 | >128 | >128 | 38.7 |
| 7 | 32 | 128 | 32 | >128 | >128 | >128 | 31.5 |
| 8 | 32 | 128 | 8 | >128 | >128 | >128 | 20.7 |
| 9 | >128 | >128 | >128 | >128 | >128 | >128 | 42.7 |
| 10 | 8 | 16 | 4 | 32 | 64 | 64 | 23.3 |
| 11 | 16 | 32 | 4 | >128 | >128 | >128 | 20.6 |
| 12 | >128 | >128 | >128 | >128 | >128 | >128 | 42.9 |
| Kanamycin | 2 | 0.06 | 0.5 | 2 | 1 | 2 | |
| Staurosporine | 0.13 | ||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shin, A.-Y.; Lee, H.-S.; Lee, J. Isolation of Scalimides A–L: β-Alanine-Bearing Scalarane Analogs from the Marine Sponge Spongia sp. Mar. Drugs 2022, 20, 726. https://doi.org/10.3390/md20110726
Shin A-Y, Lee H-S, Lee J. Isolation of Scalimides A–L: β-Alanine-Bearing Scalarane Analogs from the Marine Sponge Spongia sp. Marine Drugs. 2022; 20(11):726. https://doi.org/10.3390/md20110726
Chicago/Turabian StyleShin, A-Young, Hyi-Seung Lee, and Jihoon Lee. 2022. "Isolation of Scalimides A–L: β-Alanine-Bearing Scalarane Analogs from the Marine Sponge Spongia sp." Marine Drugs 20, no. 11: 726. https://doi.org/10.3390/md20110726
APA StyleShin, A. -Y., Lee, H. -S., & Lee, J. (2022). Isolation of Scalimides A–L: β-Alanine-Bearing Scalarane Analogs from the Marine Sponge Spongia sp. Marine Drugs, 20(11), 726. https://doi.org/10.3390/md20110726