Two New Phenylhydrazone Derivatives from the Pearl River Estuary Sediment-Derived Streptomyces sp. SCSIO 40020
Abstract
:1. Introduction
2. Results and Discussion
2.1. Structural Determination
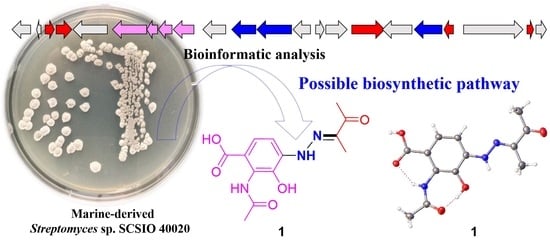
2.2. Biosynthetic Implications
2.3. Biological Activities
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Strain, Screening and Culture Methods
3.3. Extraction, Isolation and Purification
3.4. Physical and Chemical Properties of the New Compounds 1–3 and 5
3.5. X-Ray Crystallographic Analysis Data of Penzonemycin A (1) and 5
3.6. Bioactivity Assays
3.6.1. Cytotoxic Activity Assay
3.6.2. Antifungal Activity Assay
3.6.3. Antibacterial Activity Assay
3.6.4. Alpha-glucosidase Inhibition Activity
3.7. Bioinformatics Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, Z.; Kang, D.; Chen, M.; Wu, G.; Feng, D.; Zhao, T.; Zhou, Z.; Huo, Z.; Jing, L.; Zuo, X.; et al. Design, synthesis, and antiviral evaluation of novel hydrazone-substituted thiophene [3,2-d]pyrimidine derivatives as potent human immunodeficiency virus-1 inhibitors. Chem. Biol. Drug Des. 2018, 92, 2009–2021. [Google Scholar] [CrossRef] [PubMed]
- Bolognino, I.; Giangregorio, N.; Tonazzi, A.; Martínez, A.L.; Altomare, C.D.; Loza, M.I.; Sablone, S.; Cellamare, S.; Catto, M. Synthesis and biological evaluation of dantrolene-like hydrazide and hydrazone analogues as multitarget agents for neurodegenerative diseases. ChemMedChem 2021, 16, 2807–2816. [Google Scholar] [CrossRef] [PubMed]
- Achagar, R.; Elmakssoudi, A.; Dakir, M.; Elamrani, A.; Zouheir, Y.; Zahouily, M.; Jamaleddine, J. A green and efficient protocol for the synthesis of phenylhydrazone derivatives catalyzed by nanostructured diphosphate Na2CaP2O7 and screening of their antibacterial activity. ChemistrySelect 2021, 6, 1366–1371. [Google Scholar] [CrossRef]
- Ma, G.; Candra, H.; Pang, L.; Xiong, J.; Ding, Y.; Tran, H.; Low, Z.; Ye, H.; Liu, M.; Zheng, J.; et al. Biosynthesis of tasikamides via pathway coupling and diazonium-mediated hydrazone formation. J. Am. Chem. Soc. 2022, 144, 1622–1633. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Fang, S.; Shi, Z.; Wang, B.; Li, X.; Ji, N. Phenylhydrazone and quinazoline derivatives from the cold-seep-derived fungus Penicillium oxalicum. Mar. Drugs 2021, 19, 10. [Google Scholar] [CrossRef] [PubMed]
- Cheng, P.; Xu, K.; Chen, Y.; Wang, T.; Chen, Y.; Yang, C.; Ma, S.; Liang, Y.; Ge, H.; Jiao, R. Cytotoxic aromatic polyketides from an insect derived Streptomyces sp. NA4286. Tetrahedron Lett. 2019, 60, 1706–1709. [Google Scholar] [CrossRef]
- Zhang, W.; Yang, C.; Huang, C.; Zhang, L.; Zhang, H.; Zhang, Q.; Yuan, C.; Zhu, Y.; Zhang, C. Pyrazolofluostatins A–C, pyrazole-fused benzo[a]fluorenes from south China sea-derived Micromonospora rosaria SCSIO N160. Org. Lett. 2017, 19, 592–595. [Google Scholar] [CrossRef]
- Matsuda, K.; Hasebe, F.; Shiwa, Y.; Kanesaki, Y.; Tomita, T.; Yoshikawa, H.; Shin-ya, K.; Kuzuyama, T.; Nishiyama, M. Genome mining of amino group carrier protein-mediated machinery: Discovery and biosynthetic characterization of a natural product with unique hydrazone unit. ACS Chem. Biol. 2017, 12, 124–131. [Google Scholar] [CrossRef]
- Blair, L.M.; Sperry, J. Natural products containing a nitrogen–nitrogen bond. J. Nat. Prod. 2013, 76, 794–812. [Google Scholar] [CrossRef]
- Ren, X.; Xie, X.; Chen, B.; Liu, L.; Jiang, C.; Qian, Q. Marine natural products: A potential source of anti-hepatocellular carcinoma drugs. J. Med. Chem. 2021, 64, 7879–7899. [Google Scholar] [CrossRef]
- Jagannathan, S.V.; Manemann, E.M.; Rowe, S.E.; Callender, M.C.; Soto, W. Marine actinomycetes, new sources of biotechnological products. Mar. Drugs 2021, 19, 365. [Google Scholar] [CrossRef]
- Zhai, S.; Liu, W.; Wu, Z.; Zhu, Y.; Zhang, W.; Zhang, C.; Ma, L. Identification of bafilomycins and heterologous expression of its biosynthetic gene cluster from the Pearl River Estuary sediment-derived Streptomyces sp. SCSIO 40020. Acta Microbiol. Sin. 2021, 61, 3991–4005. [Google Scholar]
- Liu, N.; Song, F.; Shang, F.; Huang, Y. Mycemycins A-E, new dibenzoxazepinones isolated from two different Streptomycetes. Mar. Drugs 2015, 13, 6247–6258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, Z.; Zhao, J.; Liu, J.; Zhang, M.; Chen, R.; Xie, K.; Dai, J. Sesquiterpenoids from the cultured mycelia of Ganoderma capense. Fitoterapia 2017, 118, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Shaaban, K.A.; Singh, S.; Elshahawi, S.I.; Wang, X.; Ponomareva, L.V.; Sunkara, M.; Copley, G.C.; Hower, J.C.; Morris, A.J.; Kharel, M.K.; et al. The native production of the sesquiterpene isopterocarpolone by Streptomyces sp. RM-14-6. Nat. Prod. Res. 2014, 28, 337–339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cano-Prieto, C.; García-Salcedo, R.; Sánchez-Hidalgo, M.; Braña, A.; Fiedler, H.; Méndez, C.; Salas, J.; Olano, C. Genome mining of Streptomyces sp. Tü 6176: Characterization of the nataxazole biosynthesis pathway. Chembiochem 2015, 16, 1461–1473. [Google Scholar] [CrossRef]
- Ikeda, M. Towards bacterial strains overproducing L-tryptophan and other aromatics by metabolic engineering. Appl. Microbiol. Biotechnol. 2006, 69, 615–626. [Google Scholar] [CrossRef] [Green Version]
- Pavlikova, M.; Kamenik, Z.; Janata, J.; Kadlcik, S.; Kuzma, M.; Najmanova, L. Novel pathway of 3-hydroxyanthranilic acid formation in limazepine biosynthesis reveals evolutionary relation between phenazines and pyrrolobenzodiazepines. Sci. Rep. 2018, 8, 7810. [Google Scholar] [CrossRef]
- Gou, L.; Wu, Q.; Lin, S.; Li, X.; Liang, J.; Zhou, X.; An, D.; Deng, Z.; Wang, Z. Mutasynthesis of pyrrole spiroketal compound using calcimycin 3-hydroxy anthranilic acid biosynthetic mutant. Appl. Microbiol. Biotechnol. 2013, 97, 8183–8191. [Google Scholar] [CrossRef]
- Noguchi, T.; Isogai, S.; Terada, T.; Nishiyama, M.; Kuzuyama, T. Cryptic oxidative transamination of hydroxynaphthoquinone in natural product biosynthesis. J. Am. Chem. Soc. 2022, 144, 5435–5440. [Google Scholar] [CrossRef]
- Sugai, Y.; Katsuyama, Y.; Ohnishi, Y. A nitrous acid biosynthetic pathway for diazo group formation in bacteria. Nat. Chem. Biol. 2016, 12, 73–75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Del Rio Flores, A.; Twigg, F.F.; Du, Y.; Cai, W.; Aguirre, D.Q.; Sato, M.; Dror, M.J.; Narayanamoorthy, M.; Geng, J.; Zill, N.A.; et al. Biosynthesis of triacsin featuring an N-hydroxytriazene pharmacophore. Nat. Chem. Biol. 2021, 17, 1305–1313. [Google Scholar] [CrossRef] [PubMed]
- Chipman, D.; Barak, Z.; Schloss, J.V. Biosynthesis of 2-aceto-2-hydroxy acids: Acetolactate synthases and acetohydroxyacid synthases. Biochim. Biophys. Acta. 1998, 1385, 401–419. [Google Scholar] [CrossRef]
- Liu, W.; Zhang, W.; Jin, H.; Zhang, Q.; Chen, Y.; Jiang, X.; Zhang, G.; Zhang, L.; Zhang, W.; She, Z.; et al. Genome mining of marine-derived Streptomyces sp. SCSIO 40010 leads to cytotoxic new polycyclic tetramate macrolactams. Mar. Drugs 2019, 17, 663. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Zhang, G.; Zhang, L.; Liu, W.; Jiang, X.; Jin, H.; Liu, Z.; Zhang, H.; Zhou, A.; Zhang, C. New polycyclic tetramate macrolactams from marine-derived Streptomyces sp. SCSIO 40060. Tetrahedron 2018, 74, 6839–6845. [Google Scholar] [CrossRef]
- Dolomanov, O.V.; Bourhis, L.J.; Gildea, R.J.; Howard, J.A.K.; Puschmann, H. OLEX2: A complete structure solution, refinement and analysis program. J. Appl. Crystallogr. 2009, 42, 339–341. [Google Scholar] [CrossRef]
- Sheldrick, G.M. SHELXT—Integrated space-group and crystal-structure determination. Acta Cryst. A Found Adv. 2015, 71, 3–8. [Google Scholar] [CrossRef] [Green Version]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta. Cryst. C Struct. Chem. 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Skehan, P.; Storeng, R.; Scudiero, D.; Monks, A.; McMahon, J.; Vistica, D.; Warren, J.T.; Bokesch, H.; Kenney, S.; Boyd, M.R. New colorimetric cytotoxicity assay for anticancer-drug screening. J. Natl. Cancer. Inst. 1990, 82, 1107–1112. [Google Scholar] [CrossRef]
- Nakabayashi, H.; Taketa, K.; Miyano, K.; Yamane, T.; Sato, J. Growth of human hepatoma cells lines with differentiated functions in chemically defined medium. Cancer Res. 1982, 42, 3858–3863. [Google Scholar]
- Fang, Z.; Zhang, Q.; Zhang, L.; She, J.; Li, J.; Zhang, W.; Zhang, H.; Zhu, Y.; Zhang, C. Antifungal macrolides kongjuemycins from coral-associated rare actinomycete Pseudonocardia kongjuensis SCSIO 11457. Org. Lett. 2022, 24, 3482–3487. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Huang, C.; Fang, C.; Zhang, L.; Chen, S.; Zhang, Q.; Zhang, C.; Zhang, W. Inactivation of flavoenzyme-encoding gene flsO1 in fluostatin biosynthesis leads to diversified angucyclinone derivatives. J. Org. Chem. 2021, 86, 11019–11028. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; Van Wezel, G.P.; Medema, M.H.; Weber, T. AntiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef] [PubMed]





| Position | 1 | 2 | ||
|---|---|---|---|---|
| δC, Type | δH, Mult (J Hz) | δC, Type | δH, Mult (J Hz) | |
| 1 | 115.3, C | 117.3, C | ||
| 2 | 127.5, C | 128.0, C | ||
| 3 | 137.6, C | 136.1, C | ||
| 4 | 137.0, C | 136.4, C | ||
| 5 | 109.4, CH | 7.37, d (8.6) | 108.6, CH | 7.30, d (8.5) |
| 6 | 124.0, CH | 7.57, d (8.6) | 123.6, CH | 7.58, d (8.5) |
| 7 | 168.6, C | 169.2, C | ||
| 8-NH | 10.94, br s | 10.97, br s | ||
| 9 | 171.3, C | 174.5, C | ||
| 10 | 23.6, CH3 | 2.21, s | 29.8, CH2 | 2.49, overlapping |
| 11 | 9.8, CH3 | 1.17, t (7.6) | ||
| 12-NH | 8.94, s | 8.84, s | ||
| 14 | 144.2, C | 143.3, C | ||
| 15 | 196.3, C | 196.2, C | ||
| 16 | 8.1, CH3 | 2.00, s | 7.9, CH3 | 1.99, s |
| 17 | 24.2, CH3 | 2.40, s | 24.1, CH3 | 2.40, s |
| 3-OH | 10.18, br s | n.d. | ||
| 1-COOH | 13.00, br s | 13.09, br s | ||
| Position | 3 | |
|---|---|---|
| δC, Type | δH, Mult (J in Hz) | |
| 1 | 163.6, C | |
| 2 | 109.8, C | |
| 3 | 127.4, CH | 7.99, dd (7.7, 0.8) |
| 4 | 120.1, CH | 7.11, t (7.6) |
| 5 | 134.5, CH | 7.57, t (8.0, 1.7) |
| 6 | 117.4, CH | 7.15, d (8.3) |
| 7 | 158.4, C | |
| 8 | 149.6, C | |
| 9 | 115.4, CH | 8.12, d (8.1) |
| 10 | 125.4, CH | 7.58, t (8.0) |
| 11 | 127.5, CH | 8.05, dd (7.8, 1.5) |
| 12 | 121.8, C | |
| 13 | 138.8, C | |
| 14 | 165.6, C | |
| Gene | Size (aa) | Annotation and Closest Homologue (Source, Accession No.) | % Identity/ Similarity |
|---|---|---|---|
| pzm1 | 353 | LysR family transcriptional regulator (Streptomyces halstedii LGO-A4, MBV7672918) | 94/94 |
| pzm2 | 83 | hypothetical protein (Streptomyces halstedii LGO-A4, MBV7672919) | 100/100 |
| pzm3 | 171 | N-acetyltransferase (Streptomyces halstedii LGO-A4, MBV7672920) | 98/98 |
| pzm4 | 298 | F420-dependent oxidoreductase (Streptomyces halstedii LGO-A4, MBV7672921) | 99/99 |
| pzm5 | 653 | AfsR/SARP family transcriptional regulator (Streptomyces sp. SM18, AWL43113) | 99/99 |
| pzm6 | 650 | anthranilate synthase (Streptomyces halstedii LGO-A4, MBV7672922) | 99/99 |
| pzm7 | 207 | isochorismatase (Streptomyces halstedii LGO-A4, MBV7672923) | 99/100 |
| pzm8 | 256 | 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase (Streptomyces halstedii LGO-A4, MBV7672924) | 98/99 |
| pzm9 | 393 | 3-deoxy-d-arabino-heptulosonate 7-phosphate (DAHP) synthase(Streptomyces halstedii LGO-A4, MBV7672925) | 99/98 |
| pzm10 | 432 | transposase (Streptomyces halstedii LGO-A4, MBV7672926) | 99/99 |
| pzm11 | 459 | nitrosuccinate lyase, CreD homolog(Streptomyces cremeus NBRC 12760, BAU09301) | 67/73 |
| pzm12 | 659 | monooxygenase, CreE homolog (Streptomyces cremeus NBRC 12760, BAU09302) | 62/70 |
| pzm13 | 334 | quinone reductase (Streptomyces halstedii LGO-A4, MBV7672929) | 99/99 |
| pzm14 | 161 | Rrf2 family transcriptional regulator (Streptomyces halstedii LGO-A4, MBV7672930) | 100/100 |
| pzm15 | 406 | major facilitator superfamily (MFS) transporter (Streptomyces halstedii LGO-A4, MBV7672931) | 100/100 |
| pzm16 | 622 | acetolactate synthase (Streptomyces halstedii LGO-A4, MBV7672932) | 99/99 |
| pzm17 | 560 | Acyl-CoA ligase (Streptomyces halstedii LGO-A4, MBV7672933) | 99/99 |
| pzm18 | 527 | AMP-binding protein, CreM homolog (Streptomyces cremeus NBRC 12760, BAU09310) | 48/61 |
| pzm19 | 180 | SRPBCC family protein (Streptomyces halstedii LGO-A4, MBV7672935) | 99/99 |
| pzm20 | 1128 | discoidin domain-containing protein (Streptomyces halstedii LGO-A4, MBV7672936) | 99/99 |
| pzm21 | 126 | vicinal oxygen chelate (VOC) family protein (Streptomyces halstedii LGO-A4, MBV7672937) | 100/100 |
| pzm22 | 198 | RNA 2′,3′-cyclic phosphodiesterase (Streptomyces halstedii LGO-A4, MBV7672938) | 99/100 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, W.; Ma, L.; Zhang, L.; Chen, Y.; Zhang, Q.; Zhang, H.; Zhang, W.; Zhang, C.; Zhang, W. Two New Phenylhydrazone Derivatives from the Pearl River Estuary Sediment-Derived Streptomyces sp. SCSIO 40020. Mar. Drugs 2022, 20, 449. https://doi.org/10.3390/md20070449
Liu W, Ma L, Zhang L, Chen Y, Zhang Q, Zhang H, Zhang W, Zhang C, Zhang W. Two New Phenylhydrazone Derivatives from the Pearl River Estuary Sediment-Derived Streptomyces sp. SCSIO 40020. Marine Drugs. 2022; 20(7):449. https://doi.org/10.3390/md20070449
Chicago/Turabian StyleLiu, Wei, Liang Ma, Liping Zhang, Yuchan Chen, Qingbo Zhang, Haibo Zhang, Weimin Zhang, Changsheng Zhang, and Wenjun Zhang. 2022. "Two New Phenylhydrazone Derivatives from the Pearl River Estuary Sediment-Derived Streptomyces sp. SCSIO 40020" Marine Drugs 20, no. 7: 449. https://doi.org/10.3390/md20070449
APA StyleLiu, W., Ma, L., Zhang, L., Chen, Y., Zhang, Q., Zhang, H., Zhang, W., Zhang, C., & Zhang, W. (2022). Two New Phenylhydrazone Derivatives from the Pearl River Estuary Sediment-Derived Streptomyces sp. SCSIO 40020. Marine Drugs, 20(7), 449. https://doi.org/10.3390/md20070449