Eighteen Novel Bioactive Peptides from Monkfish (Lophius litulon) Swim Bladders: Production, Identification, Antioxidant Activity, and Stability
Abstract
:1. Introduction
2. Results
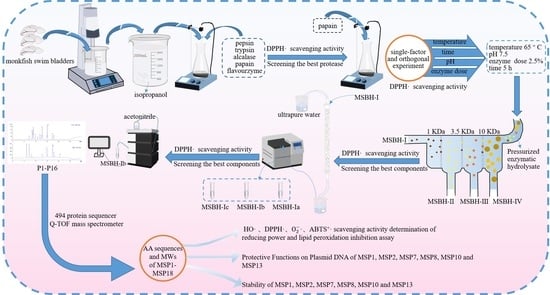
2.1. Preparation of Protein Hydrolysate of Monkfish Swim Bladders (MSBH)
2.1.1. Screening of Enzyme Species
2.1.2. Optimization of Hydrolysis Conditions of Papain
2.2. Preparation of APs from MSBH
2.2.1. Ultrafiltration
2.2.2. Chromatography of MSBH
2.3. Determination of the AA Sequences and MWs of Eighteen Isolated APs (MSP1-MSP18)
2.4. Antioxidant Activity of MSP1 to MSP18
2.4.1. Radical Scavenging Activity of Eighteen Isolated APs (MSP1-MSP18)
2.4.2. Lipid Peroxidation Inhibition Ability
2.4.3. Ferric Reducing Antioxidant Power (FRAP)
2.4.4. Protective Effect on H2O2-Damaged Plasmid DNA
2.4.5. Cytoprotective Function on H2O2-Damaged HepG2 Cells
2.5. Stability of MSP1, MSP2, MSP7, MSP8, MSP10 and MSP13
2.5.1. pH Stability of MSP1, MSP2, MSP7, MSP8, MSP10 and MSP13
2.5.2. Thermal Stability of MSP1, MSP2, MSP7, MSP8, MSP10 and MSP13
2.5.3. Stability of MSP1, MSP2, MSP7, MSP8, MSP10 and MSP13 Subjected to Simulated Gastrointestinal (GI) Digestion
3. Discussion
3.1. Preparation of APs from MSBH
3.2. Antioxidant Activity of MSP1, MSP2, MSP7, MSP8, MSP10 and MSP13
3.3. Structure–Activity Relationship of MSP1, MSP2, MSP7, MSP8, MSP10 and MSP13
4. Materials and Methods
4.1. Materials and Chemical Reagents
4.2. Preparation of MSBH
4.2.1. Screening of Enzyme Species
4.2.2. Optimization of Hydrolysis Conditions of Papain
4.3. Preparation of APs from MSBH
4.3.1. Ultrafiltration of MSBH
4.3.2. Purification of APs from MSBH-I by Chromatography Methods
4.4. Identification of Eighteen Isolated APs (MSP1-MSP18)
4.5. Antioxidant Activity of MSP1 to MSP18
4.5.1. HO· Scavenging Activity
4.5.2. DPPH· Scavenging Activity
4.5.3. Scavenging Activity
4.5.4. ABTS+· Scavenging Activity
4.5.5. Determination of Reducing Power
4.5.6. Lipid Peroxidation Inhibition Assay
4.5.7. Protective Functions on Plasmid DNA of MSP1, MSP2, MSP7, MSP8, MSP10 and MSP13
4.5.8. Cytoprotection of MSP1, MSP2, MSP7, MSP8, MSP10 and MSP13 on H2O2-Damaged HepG2 Cells
4.6. Stability of MSP1, MSP2, MSP7, MSP8, MSP10 and MSP13
4.7. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Ahmed, M.; Verma, A.K.; Patel, R. Collagen extraction and recent biological activities of collagen peptides derived from sea-food waste: A review. Sustain. Chem. Pharm. 2020, 18, 100315. [Google Scholar] [CrossRef]
- Zhang, L.; Zhao, G.-X.; Zhao, Y.-Q.; Qiu, Y.-T.; Chi, C.-F.; Wang, B. Identification and Active Evaluation of Antioxidant Peptides from Protein Hydrolysates of Skipjack Tuna (Katsuwonus pelamis) Head. Antioxidants 2019, 8, 318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wen, C.; Zhang, J.; Zhang, H.; Duan, Y.; Ma, H. Plant protein-derived antioxidant peptides: Isolation, identification, mechanism of action and application in food systems: A review. Trends Food Sci. Technol. 2020, 105, 308–322. [Google Scholar] [CrossRef]
- Chakrabarti, S.; Guha, S.; Majumder, K. Food-Derived Bioactive Peptides in Human Health: Challenges and Opportunities. Nutrients 2018, 10, 1738. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.-Y.; Zhao, Y.-Q.; Wang, Y.-M.; Yang, X.-R.; Chi, C.-F.; Wang, B. Gelatins and antioxidant peptides from Skipjack tuna (Katsuwonus pelamis) skins: Purification, characterization, and cytoprotection on ultraviolet—A injured human skin fibroblasts. Food Biosci. 2022, 50, 102138. [Google Scholar] [CrossRef]
- Qiao, Q.-Q.; Luo, Q.-B.; Suo, S.-K.; Zhao, Y.-Q.; Chi, C.-F.; Wang, B. Preparation, Characterization, and Cytoprotective Effects on HUVECs of Fourteen Novel Angiotensin-I-Converting Enzyme Inhibitory Peptides From Protein Hydrolysate of Tuna Processing By-Products. Front. Nutr. 2022, 9, 868671. [Google Scholar] [CrossRef]
- Admassu, H.; Gasmalla, M.A.A.; Yang, R.; Zhao, W. Bioactive Peptides Derived from Seaweed Protein and Their Health Benefits: Antihypertensive, Antioxidant, and Antidiabetic Properties. J. Food Sci. 2018, 83, 6–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdelhedi, O.; Nasri, M. Basic and recent advances in marine antihypertensive peptides: Production, structure-activity relationship and bioavailability. Trends Food Sci. Technol. 2019, 88, 543–557. [Google Scholar] [CrossRef]
- Mardani, M.; Badakné, K.; Farmani, J.; Aluko, R.E. Antioxidant peptides: Overview of production, properties, and applications in food systems. Compr. Rev. Food Sci. Food Saf. 2023, 22, 46–106. [Google Scholar] [CrossRef] [PubMed]
- Ren, L.-K.; Yang, Y.; Ma, C.-M.; Fan, J.; Bian, X.; Liu, B.-X.; Wang, D.-F.; Zhu, P.-Y.; Fu, Y.; Zhang, N. Identification and in silico analysis of novel antioxidant peptides in broken rice protein hydrolysate and its cytoprotective effect against H2O2-induced 2BS cell model. Food Res. Int. 2022, 162, 112108. [Google Scholar] [CrossRef]
- Pan, X.-Y.; Wang, Y.-M.; Li, L.; Chi, C.-F.; Wang, B. Four Antioxidant Peptides from Protein Hydrolysate of Red Stingray (Dasyatis akajei) Cartilages: Isolation, Identification, and In Vitro Activity Evaluation. Mar. Drugs 2019, 17, 263. [Google Scholar] [CrossRef] [Green Version]
- Xiang, Z.; Xue, Q.; Gao, P.; Yu, H.; Wu, M.; Zhao, Z.; Li, Y.; Wang, S.; Zhang, J.; Dai, L. Antioxidant peptides from edible aquatic animals: Preparation method, mechanism of action, and structure-activity relationships. Food Chem. 2023, 404, 134701. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Wang, Y.-M.; Chi, C.-F.; Luo, H.-Y.; Deng, S.-G.; Ma, J.-Y. Isolation and Characterization of Collagen and Antioxidant Collagen Peptides from Scales of Croceine Croaker (Pseudosciaena crocea). Mar. Drugs 2013, 11, 4641–4661. [Google Scholar] [CrossRef]
- Zhu, L.; Xiong, H.; Huang, X.; Guyonnet, V.; Ma, M.; Chen, X.; Zheng, Y.; Wang, L.; Hu, G. Identification and molecular mechanisms of novel antioxidant peptides from two sources of eggshell membrane hydrolysates showing cytoprotection against oxidative stress: A combined in silico and in vitro study. Food Res. Int. 2022, 157, 111266. [Google Scholar] [CrossRef] [PubMed]
- Dresen, E.; Pimiento, J.M.; Patel, J.J.; Heyland, D.K.; Rice, T.W.; Stoppe, C. Overview of oxidative stress and the role of micronutrients in critical illness. J. Parenter. Enter. Nutr. 2022, 47, S38–S39. [Google Scholar] [CrossRef] [PubMed]
- García, N.; Zazueta, C.; Aguilera-Aguirre, L. Oxidative Stress and Inflammation in Cardiovascular Disease. Oxid. Med. Cell. Longev. 2017, 2017, 5853238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, Y.R.; Park, C.-I.; Soh, Y. Antioxidant and Anti-Inflammatory Effects of NCW Peptide from Clam Worm (Marphysa sanguinea). J. Microbiol. Biotechnol. 2020, 30, 1387–1394. [Google Scholar] [CrossRef]
- Hu, X.-M.; Wang, Y.-M.; Zhao, Y.-Q.; Chi, C.-F.; Wang, B. Antioxidant Peptides from the Protein Hydrolysate of Monkfish (Lophius litulon) Muscle: Purification, Identification, and Cytoprotective Function on HepG2 Cells Damage by H2O2. Mar. Drugs 2020, 18, 153. [Google Scholar] [CrossRef] [Green Version]
- Forman, H.J.; Zhang, H. Targeting oxidative stress in disease: Promise and limitations of antioxidant therapy. Nat. Rev. Drug Discov. 2021, 20, 689–709. [Google Scholar] [CrossRef]
- Sheng, Y.; Qiu, Y.T.; Wang, Y.M.; Chi, C.F.; Wang, B. Novel antioxidant collagen peptides of siberian sturgeon (Acipenser baerii) cartilages: The preparation, characterization, and cytoprotection of H2O2-damaged human umbilical vein endothelial cells (HUVECs). Mar. Drugs 2022, 20, 325. [Google Scholar] [CrossRef]
- Kong, J.; Hu, X.-M.; Cai, W.-W.; Wang, Y.-M.; Chi, C.-F.; Wang, B. Bioactive Peptides from Skipjack Tuna Cardiac Arterial Bulbs (II): Protective Function on UVB-Irradiated HaCaT Cells through Antioxidant and Anti-Apoptotic Mechanisms. Mar. Drugs 2023, 20, 105. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Li, T.; Chen, D.; Gu, H.; Mao, X. Identification and molecular docking of antioxidant peptides from hemp seed protein hydrolysates. LWT 2021, 147, 111453. [Google Scholar] [CrossRef]
- Wong, F.-C.; Xiao, J.; Wang, S.; Ee, K.-Y.; Chai, T.-T. Advances on the antioxidant peptides from edible plant sources. Trends Food Sci. Technol. 2020, 99, 44–57. [Google Scholar] [CrossRef]
- Wang, Y.-Z.; Wang, Y.-M.; Pan, X.; Chi, C.-F.; Wang, B. Antioxidant Mechanisms of the Oligopeptides (FWKVV and FMPLH) from Muscle Hydrolysate of Miiuy Croaker against Oxidative Damage of HUVECs. Oxid. Med. Cell. Longev. 2021, 2021, 9987844. [Google Scholar] [CrossRef] [PubMed]
- Sila, A.; Bougatef, A. Antioxidant peptides from marine by-products: Isolation, identification and application in food systems. A review. J. Funct. Foods 2016, 21, 10–26. [Google Scholar] [CrossRef]
- Wang, Y.-Z.; Zhao, Y.-Q.; Wang, Y.-M.; Zhao, W.-H.; Wang, P.; Chi, C.-F.; Wang, B. Antioxidant peptides from Antarctic Krill (Euphausia superba) hydrolysate: Preparation, identification and cytoprotection on H2O2-induced oxidative stress. J. Funct. Foods 2021, 86, 104701. [Google Scholar] [CrossRef]
- Achar, J.C.; Nam, G.; Jung, J.; Klammler, H.; Mohamed, M.M. Microbubble ozonation of the antioxidant butylated hydroxytoluene: Degradation kinetics and toxicity reduction. Environ. Res. 2020, 186, 109496. [Google Scholar] [CrossRef]
- Liang, X.; Zhao, Y.; Liu, W.; Li, Z.; Souders, C.L.; Martyniuk, C.J. Butylated hydroxytoluene induces hyperactivity and alters dopamine-related gene expression in larval zebrafish (Danio rerio). Environ. Pollut. 2020, 257, 113624. [Google Scholar] [CrossRef]
- Eskandani, M.; Hamishehkar, H.; Dolatabadi, J.E.N. Cytotoxicity and DNA damage properties of tert-butylhydroquinone (TBHQ) food additive. Food Chem. 2014, 153, 315–320. [Google Scholar] [CrossRef]
- Botterweck, A.; Verhagen, H.; Goldbohm, R.; Kleinjans, J.; Brandt, P.V.D. Intake of butylated hydroxyanisole and butylated hydroxytoluene and stomach cancer risk: Results from analyses in the Netherlands Cohort Study. Food Chem. Toxicol. 2000, 38, 599–605. [Google Scholar] [CrossRef] [Green Version]
- Sun, K.-L.; Gao, M.; Wang, Y.-Z.; Li, X.-R.; Wang, P.; Wang, B. Antioxidant Peptides From Protein Hydrolysate of Marine Red Algae Eucheuma cottonii: Preparation, Identification, and Cytoprotective Mechanisms on H2O2 Oxidative Damaged HUVECs. Front. Microbiol. 2022, 13, 791248. [Google Scholar] [CrossRef]
- Ahn, C.-B.; Je, J.-Y.; Cho, Y.-S. Antioxidant and anti-inflammatory peptide fraction from salmon byproduct protein hydrolysates by peptic hydrolysis. Food Res. Int. 2012, 49, 92–98. [Google Scholar] [CrossRef]
- Senphan, T.; Benjakul, S. Antioxidative activities of hydrolysates from seabass skin prepared using protease from hepatopancreas of Pacific white shrimp. J. Funct. Foods 2014, 6, 147–156. [Google Scholar] [CrossRef]
- Chi, C.-F.; Wang, B.; Hu, F.-Y.; Wang, Y.-M.; Zhang, B.; Deng, S.-G.; Wu, C.-W. Purification and identification of three novel antioxidant peptides from protein hydrolysate of bluefin leatherjacket (Navodon septentrionalis) skin. Food Res. Int. 2015, 73, 124–129. [Google Scholar] [CrossRef]
- Leo, E.E.M.; Fernández, J.J.A.; Campos, M.R.S. Biopeptides with antioxidant and anti-inflammatory potential in the prevention and treatment of diabesity disease. Biomed. Pharmacother. 2016, 83, 816–826. [Google Scholar] [CrossRef]
- Landim, A.P.M.; Tiburski, J.H.; Mellinger, C.G.; Juliano, P.; Rosenthal, A. Potential Application of High Hydrostatic Pressure on the Production of Hydrolyzed Proteins with Antioxidant and Antihypertensive Properties and Low Allergenicity: A Review. Foods 2023, 12, 630. [Google Scholar] [CrossRef]
- Wang, Y.-M.; Pan, X.; He, Y.; Chi, C.-F.; Wang, B. Hypolipidemic Activities of Two Pentapeptides (VIAPW and IRWWW) from Miiuy Croaker (Miichthys miiuy) Muscle on Lipid Accumulation in HepG2 Cells through Regulation of AMPK Pathway. Appl. Sci. 2020, 10, 817. [Google Scholar] [CrossRef] [Green Version]
- Ma, R.; Chen, Q.; Dai, Y.; Huang, Y.; Hou, Q.; Huang, Y.; Zhong, K.; Huang, Y.; Gao, H.; Bu, Q. Identification of novel antioxidant peptides from sea squirt (Halocynthia roretzi) and its neuroprotective effect in 6-OHDA-induced neurotoxicity. Food Funct. 2022, 13, 6008–6021. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yu, H.; Xing, R.; Li, P. Hepatoprotective Effect of Oyster Peptide on Alcohol-Induced Liver Disease in Mice. Int. J. Mol. Sci. 2022, 23, 8081. [Google Scholar] [CrossRef]
- Jiang, Y.; Wang, R.; Yin, Z.; Sun, J.; Wang, B.; Zhao, D.; Zeng, X.A.; Li, H.; Huang, M.; Sun, B. Optimization of Jiuzao protein hydrolysis conditions and antioxidant activity in vivo of Jiuzao tetrapeptide Asp-Arg-Glu-Leu by elevating the Nrf2/Keap1-p38/PI3K-MafK signaling pathway. Food Funct. 2021, 12, 4808–4824. [Google Scholar] [CrossRef]
- Chi, C.-F.; Wang, B.; Deng, Y.-Y.; Wang, Y.-M.; Deng, S.-G.; Ma, J.-Y. Isolation and characterization of three antioxidant pentapeptides from protein hydrolysate of monkfish (Lophius litulon) muscle. Food Res. Int. 2014, 55, 222–228. [Google Scholar] [CrossRef]
- Tian, X.; Zheng, J.; Xu, B.; Ye, J.; Yang, Z.; Yuan, F. Optimization of extraction of bioactive peptides from monkfish (Lophius litulon) and characterization of their role in H2O2-Induced Lesion. Mar. Drugs 2020, 18, 468. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Tian, X.; Wang, Q.; Zheng, J.; Yang, Y.; Xu, B.; Zhang, S.; Yuan, F.; Yang, Z. Monkfish Peptides Mitigate High Fat Diet-Induced Hepatic Steatosis in Mice. Mar. Drugs 2022, 20, 312. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Miao, B.; Cao, H.; Tian, X.; Shen, L.; Yang, Z.; Yuan, F.; Ding, Y. Monkfish (Lophius litulon) Peptides Ameliorate High-Fat-Diet-Induced Nephrotoxicity by Reducing Oxidative Stress and Inflammation via Regulation of Intestinal Flora. Molecules 2022, 28, 245. [Google Scholar] [CrossRef]
- Ren, Z.; Yang, F.; Yao, S.; Bi, L.; Jiang, G.; Huang, J.; Tang, Y. Effects of low molecular weight peptides from monkfish (Lophius litulon) roe on immune response in immunosuppressed mice. Front. Nutr. 2022, 9, 929105. [Google Scholar] [CrossRef]
- Miao, B.; Zheng, J.; Zheng, G.; Tian, X.; Zhang, W.; Yuan, F.; Yang, Z. Using Collagen Peptides From the Skin of Monkfish (Lophius litulon) to Ameliorate Kidney Damage in High-Fat Diet Fed Mice by Regulating the Nrf2 Pathway and NLRP3 Signaling. Front. Nutr. 2022, 9, 798708. [Google Scholar] [CrossRef]
- Zhang, S.Y.; Zhao, G.X.; Suo, S.K.; Wang, Y.M.; Chi, C.F.; Wang, B. Purification, identification, activity evaluation, and stability of antioxidant peptides from Alcalase hydrolysate of Antarctic Krill (Euphausia superba) proteins. Mar. Drugs 2021, 19, 347. [Google Scholar] [CrossRef]
- Khammuang, S.; Sarnthima, R.; Sanachai, K. Purification and identification of novel antioxidant peptides from silkworm pupae (Bombyx mori) protein hydrolysate and molecular docking study. Biocatal. Agric. Biotechnol. 2022, 42, 102367. [Google Scholar] [CrossRef]
- Zhang, J.B.; Zhao, Y.Q.; Wang, Y.M.; Chi, C.F.; Wang, B. Eight peptides from collagen hydrolysate fraction of Spanish mackerel (Scomberomorous niphonius) skin: Isolation, identification, and antioxidant activity in vitro. Mar. Drugs 2019, 17, 224. [Google Scholar] [CrossRef] [Green Version]
- Ahn, C.-B.; Lee, K.-H.; Je, J.-Y. Enzymatic production of bioactive protein hydrolysates from tuna liver: Effects of enzymes and molecular weight on bioactivity. Int. J. Food Sci. Technol. 2010, 45, 562–568. [Google Scholar] [CrossRef]
- Jang, H.L.; Shin, S.R.; Yoon, K.Y. Hydrolysis conditions for antioxidant peptides derived from enzymatic hydrolysates of sandfish (Arctoscopus japonicus). Food Sci. Biotechnol. 2017, 26, 1191–1197. [Google Scholar] [CrossRef] [PubMed]
- Chi, C.-F.; Wang, B.; Wang, Y.-M.; Zhang, B.; Deng, S.-G. Isolation and characterization of three antioxidant peptides from protein hydrolysate of bluefin leatherjacket (Navodon septentrionalis) heads. J. Funct. Foods 2015, 12, 1–10. [Google Scholar] [CrossRef]
- Kim, H.-S.; Je, J.-G.; Ryu, B.; Kang, N.; Fernando, I.P.S.; Jayawardena, T.U.; Sanjeewa, K.K.A.; Oh, J.-Y.; Lee, T.-G.; Jeon, Y.-J. Antioxidant and angiotensin-I converting enzyme inhibitory peptides from Hippocampus abdominalis. Eur. Food Res. Technol. 2019, 245, 479–487. [Google Scholar] [CrossRef]
- Xia, Z.; Miao, J.; Chen, B.; Guo, J.; Ou, Y.; Liang, X.; Yin, Y.; Tong, X.; Cao, Y. Purification, identification, and antioxidative mechanism of three novel selenium-enriched oyster antioxidant peptides. Food Res. Int. 2022, 157, 111359. [Google Scholar] [CrossRef]
- Wang, B.; Li, Z.-R.; Chi, C.-F.; Zhang, Q.-H.; Luo, H.-Y. Preparation and evaluation of antioxidant peptides from ethanol-soluble proteins hydrolysate of Sphyrna lewini muscle. Peptides 2012, 36, 240–250. [Google Scholar] [CrossRef]
- Luo, H.-Y.; Wang, B.; Li, Z.-R.; Chi, C.-F.; Zhang, Q.-H.; He, G.-Y. Preparation and evaluation of antioxidant peptide from papain hydrolysate of Sphyrna lewini muscle protein. LWT Food Sci. Technol. 2013, 51, 281–288. [Google Scholar] [CrossRef]
- Cai, W.-W.; Hu, X.-M.; Wang, Y.-M.; Chi, C.-F.; Wang, B. Bioactive Peptides from Skipjack Tuna Cardiac Arterial Bulbs: Preparation, Identification, Antioxidant Activity, and Stability against Thermal, pH, and Simulated Gastrointestinal Digestion Treatments. Mar. Drugs 2022, 20, 626. [Google Scholar] [CrossRef]
- Wang, J.; Wang, Y.-M.; Li, L.-Y.; Chi, C.-F.; Wang, B. Twelve Antioxidant Peptides From Protein Hydrolysate of Skipjack Tuna (Katsuwonus pelamis) Roe Prepared by Flavourzyme: Purification, Sequence Identification, and Activity Evaluation. Front. Nutr. 2022, 8, 813780. [Google Scholar] [CrossRef] [PubMed]
- You, L.; Zhao, M.; Regenstein, J.M.; Ren, J. Purification and identification of antioxidative peptides from loach (Misgurnus anguillicaudatus) protein hydrolysate by consecutive chromatography and electrospray ionization-mass spectrometry. Food Res. Int. 2010, 43, 1167–1173. [Google Scholar] [CrossRef]
- Cai, L.; Wu, X.; Zhang, Y.; Li, X.; Ma, S.; Li, J. Purification and characterization of three antioxidant peptides from protein hydrolysate of grass carp (Ctenopharyngodon idella) skin. J. Funct. Foods 2015, 16, 234–242. [Google Scholar] [CrossRef]
- Wang, Y.-M.; Li, X.-Y.; Wang, J.; He, Y.; Chi, C.-F.; Wang, B. Antioxidant peptides from protein hydrolysate of skipjack tuna milt: Purification, identification, and cytoprotection on H2O2 damaged human umbilical vein endothelial cells. Process Biochem. 2022, 113, 258–269. [Google Scholar] [CrossRef]
- Ayala, A.; Muñoz, M.F.; Argüelles, S. Lipid peroxidation: Production, metabolism, and signaling mechanisms of malondialdehyde and 4-hydroxy-2-nonenal. Oxid. Med. Cell. Longev. 2014, 2014, 360438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Wang, B.; Chi, C.; Luo, H.; Gong, Y.; Ding, G. Influence of average molecular weight on antioxidant and functional properties of collagen hydrolysates from Sphyrna lewini, Dasyatis akjei and Raja porosa. Food Res. Int. 2013, 51, 283–293. [Google Scholar] [CrossRef]
- He, Y.; Pan, X.; Chi, C.-F.; Sun, K.-L.; Wang, B. Ten new pentapeptides from protein hydrolysate of miiuy croaker (Miichthys miiuy) muscle: Preparation, identification, and antioxidant activity evaluation. LWT 2019, 105, 1–8. [Google Scholar] [CrossRef]
- Yang, X.-R.; Zhao, Y.-Q.; Qiu, Y.-T.; Chi, C.-F.; Wang, B. Preparation and Characterization of Gelatin and Antioxidant Peptides from Gelatin Hydrolysate of Skipjack Tuna (Katsuwonus pelamis) Bone Stimulated by in vitro Gastrointestinal Digestion. Mar. Drugs 2019, 17, 78. [Google Scholar] [CrossRef] [Green Version]
- Ktari, N.; Fakhfakh, N.; Balti, R.; Ben Khaled, H.; Nasri, M.; Bougatef, A. Effect of Degree of Hydrolysis and Protease Type on the Antioxidant Activity of Protein Hydrolysates From Cuttlefish (Sepia officinalis) By-Products. J. Aquat. Food Prod. Technol. 2013, 22, 436–448. [Google Scholar] [CrossRef]
- Wang, B.; Li, L.; Chi, C.F.; Ma, J.H.; Luo, H.Y.; Xu, Y.F. Purification and characterization of a novel antioxidant peptide derived from blue mussel (Mytilus edulis) protein hydrolysate. Food Chem. 2013, 138, 1713–1719. [Google Scholar] [CrossRef]
- Bougatef, A.; Hajji, M.; Balti, R.; Lassoued, I.; Triki-Ellouz, Y.; Nasri, M. Antioxidant and free radical-scavenging activities of smooth hound (Mustelus mustelus) muscle protein hydrolysates obtained by gastrointestinal proteases. Food Chem. 2009, 114, 1198–1205. [Google Scholar] [CrossRef]
- Zou, T.-B.; He, T.-P.; Li, H.-B.; Tang, H.-W.; Xia, E.-Q. The Structure-Activity Relationship of the Antioxidant Peptides from Natural Proteins. Molecules 2016, 21, 72. [Google Scholar] [CrossRef]
- Rahman, M.S.; Hee, C.Y.; Seok, C.Y.; Alam, M.B.; Han, L.S.; Cheol, Y.J. A novel antioxidant peptide, purified from Bacillus amyloliquefaciens, showed strong antioxidant potential via Nrf-2 mediated heme oxygenase-1 expression. Food Chem. 2018, 239, 502–510. [Google Scholar] [CrossRef]
- Tonolo, F.; Moretto, L.; Grinzato, A.; Fiorese, F.; Folda, A.; Scalcon, V.; Ferro, S.; Arrigoni, G.; Bellamio, M.; Feller, E.; et al. Fermented soy-derived bioactive peptides selected by a molecular docking approach show antioxidant properties involving the Keap1/Nrf2 Pathway. Antioxidants 2020, 9, 1306. [Google Scholar] [CrossRef]
- Sanjukta, S.; Padhi, S.; Sarkar, P.; Singh, S.P.; Sahoo, D.; Rai, A.K. Production, characterization and molecular docking of antioxidant peptides from peptidome of kinema fermented with proteolytic Bacillus spp. Food Res. Int. 2021, 141, 110161. [Google Scholar] [CrossRef]
- Zhu, Y.; Lao, F.; Pan, X.; Wu, J. Food Protein-Derived Antioxidant Peptides: Molecular Mechanism, Stability and Bioavailability. Biomolecules 2022, 12, 1622. [Google Scholar] [CrossRef]
- Sheih, I.-C.; Wu, T.-K.; Fang, T.J. Antioxidant properties of a new antioxidative peptide from algae protein waste hydrolysate in different oxidation systems. Bioresour. Technol. 2009, 100, 3419–3425. [Google Scholar] [CrossRef]
- Chang, O.K.; Ha, G.E.; Han, G.-S.; Seol, K.-H.; Kim, H.W.; Jeong, S.-G.; Oh, M.-H.; Park, B.-Y.; Ham, J.-S. Novel Antioxidant Peptide Derived from the Ultrafiltrate of Ovomucin Hydrolysate. J. Agric. Food Chem. 2013, 61, 7294–7300. [Google Scholar] [CrossRef]
- Phongthai, S.; D’Amico, S.; Schoenlechner, R.; Homthawornchoo, W.; Rawdkuen, S. Fractionation and antioxidant properties of rice bran protein hydrolysates stimulated by in vitro gastrointestinal digestion. Food Chem. 2018, 240, 156–164. [Google Scholar] [CrossRef]
- Memarpoor-Yazdi, M.; Asoodeh, A.; Chamani, J. A novel antioxidant and antimicrobial peptide from hen egg white lysozyme hydrolysates. J. Funct. Foods 2012, 4, 278–286. [Google Scholar] [CrossRef]
- Zheng, Z.; Si, D.; Ahmad, B.; Li, Z.; Zhang, R. A novel antioxidative peptide derived from chicken blood corpuscle hydrolysate. Food Res. Int. 2018, 106, 410–419. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.-H.; Luo, Q.-B.; Pan, X.; Chi, C.-F.; Sun, K.-L.; Wang, B. Preparation, identification, and activity evaluation of ten antioxidant peptides from protein hydrolysate of swim bladders of miiuy croaker (Miichthys miiuy). J. Funct. Foods 2018, 47, 503–511. [Google Scholar] [CrossRef]
- Suo, S.K.; Zhao, Y.Q.; Wang, Y.M.; Pan, X.Y.; Chi, C.F.; Wang, B. Seventeen novel angiotensin converting enzyme (ACE) inhibitory peptides from protein hydrolysate of Mytilus edulis: Isolation, identification, molecular docking study, and protective function on HUVECs. Food Funct. 2022, 13, 7831–7846. [Google Scholar] [CrossRef] [PubMed]
- Suo, S.K.; Zheng, S.L.; Chi, C.F.; Luo, H.Y.; Wang, B. Novel ACE inhibitory peptides from tuna byproducts-milts: Preparation, characterization, molecular docking study and antioxidant function on H2O2-damaged HUVECs. Front. Nutr. 2022, 9, 957778. [Google Scholar] [CrossRef]
- Zheng, S.-L.; Luo, Q.-B.; Suo, S.-K.; Zhao, Y.-Q.; Chi, C.-F.; Wang, B. Preparation, identification, molecular docking study and protective function on HUVECs of novel ACE inhibitory peptides from protein hydrolysate of Skipjack tuna muscle. Mar. Drugs 2022, 20, 176. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Liu, Y.; Zhang, T.; Lv, C.; Zang, J.; Zhao, G. Structural comparison between the DNA-protective ability of scallop and shrimp ferritin from iron-induced oxidative damage. Food Chem. 2022, 386, 132827. [Google Scholar] [CrossRef] [PubMed]
- Sahoo, S.; Rath, D.; Kar, D.M.; Pattanaik, S. Hepatoprotective potency of Litsea glutinosa (L.) C.B. Rob. leaf methanol extract on H2O2-induced toxicity in HepG2 cells. J. Ethnopharmacol. 2023, 304, 116076. [Google Scholar] [CrossRef]
- Salla, S.; Sunkara, R.; Ogutu, S.; Walker, L.T.; Verghese, M. Antioxidant activity of papaya seed extracts against H2O2 induced oxidative stress in HepG2 cells. LWT 2016, 66, 293–297. [Google Scholar] [CrossRef]
- Wang, W.-Y.; Zhao, Y.-Q.; Zhao, G.-X.; Chi, C.-F.; Wang, B. Antioxidant peptides from collagen hydrolysate of Redlip croaker (Pseudosciaena polyactis) scales: Preparation, characterization, and cytoprotective effects on H2O2-damaged HepG2 cells. Mar. Drugs 2020, 18, 156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, K.; Luo, Q.; Hong, H.; Liu, H.; Luo, Y. Novel antioxidant and ACE inhibitory peptide identified from Arthrospira platensis protein and stability against thermal/pH treatments and simulated gastrointestinal digestion. Food Res. Int. 2020, 139, 109908. [Google Scholar] [CrossRef]
- Wong, F.-C.; Xiao, J.; Ong, M.G.-L.; Pang, M.-J.; Wong, S.-J.; Teh, L.K.; Chai, T.-T. Identification and characterization of antioxidant peptides from hydrolysate of blue-spotted stingray and their stability against thermal, pH and simulated gastrointestinal digestion treatments. Food Chem. 2019, 271, 614–622. [Google Scholar] [CrossRef]
- Zhao, G.-X.; Yang, X.-R.; Wang, Y.-M.; Zhao, Y.-Q.; Chi, C.-F.; Wang, B. Antioxidant peptides from the protein hydrolysate of spanish mackerel (Scomberomorous niphonius) muscle by in vitro gastrointestinal digestion and their in vitro activities. Mar. Drugs 2019, 17, 531. [Google Scholar] [CrossRef] [Green Version]












| No | Factors | DPPH· Scavenging Activity (%) | |||
|---|---|---|---|---|---|
| A Temperature (°C) | B pH | C Enzyme Dose (%) | D Time (h) | ||
| 1 | 1 (60) | 1 (7.0) | 1 (1.5) | 1 (3) | 38.58 ± 1.39 |
| 2 | 1 (60) | 2 (7.5) | 2 (2.0) | 2 (4) | 43.33 ± 0.92 |
| 3 | 1 (60) | 3 (8.0) | 3 (2.5) | 3 (5) | 43.00 ± 0.38 |
| 4 | 2 (65) | 1 (7.0) | 2 (2.0) | 3 (5) | 43.47 ± 0.45 |
| 5 | 2 (65) | 2 (7.5) | 3 (2.5) | 1 (3) | 44.63 ± 0.65 |
| 6 | 2 (65) | 3 (8.0) | 1 (1.5) | 2 (4) | 41.59 ± 1.05 |
| 7 | 3 (70) | 1 (7.0) | 3 (2.5) | 2 (4) | 44.08 ± 0.87 |
| 8 | 3 (70) | 2 (7.5) | 1 (1.5) | 3 (5) | 42.67 ± 2.52 |
| 9 | 3 (70) | 3 (8.0) | 2 (2.0) | 1 (3) | 38.67 ± 0.52 |
| K1 | 124.91 | 126.13 | 122.84 | 121.88 | |
| K2 | 129.69 | 130.63 | 125.47 | 129.00 | |
| K3 | 125.42 | 123.26 | 131.71 | 129.14 | |
| k1 | 41.64 | 42.04 | 40.95 | 40.63 | |
| k2 | 43.23 | 43.54 | 41.82 | 43.00 | |
| k3 | 41.81 | 41.09 | 43.90 | 43.05 | |
| Best level | A2 | B2 | C3 | D3 | |
| R | 1.59 | 2.45 | 2.95 | 2.42 | |
| R order | C > B > D > A | ||||
| Peaks | RT (min) | AA Sequence | Theoretical/Observed MW (Da) |
|---|---|---|---|
| P1 | 6.08 | MSP1: Tyr-Asp-Tyr-Asp (YDYD) | 574.54/574.54 |
| MSP2: Gln-Asp-Tyr-Asp (QDYD) | 539.49/539.49 | ||
| P2 | 7.14 | MSP3: Ala-Gly-Pro-Ala-Ser (AGPAS) | 401.41/401.41 |
| P3 | 8.22 | MSP4: Gly-Pro-Gly-Pro-His-Gly- Pro-Ser-Gly-Pro (GPGPHGPSGP) | 858.90/858.89 |
| P4 | 8.30 | MSP5: Gly-Pro-Lys (GPK) | 300.35/300.35 |
| P5 | 8.93 | MSP6: His-Arg-Glu (HRE) | 440.45/440.45 |
| P6 | 9.02 | MSP7: Gly-Arg-Trp (GRW) | 417.46/417.46 |
| P7 | 9.87 | MSP8: Ala-Arg-Trp (ARW) | 431.49/431.49 |
| P8 | 10.89 | MSP9: Gly-Pro-Thr-Glu (GPTE) | 402.40/402.40 |
| P9 | 10.91 | MSP 10: Asp-Asp-Gly-Gly-Lys (DDGGK) | 490.47/490.47 |
| P10 | 12.60 | MSP11: Ile-Gly-Pro-Ala-Ser (IGPAS) | 443.49/443.49 |
| MSP12: Ala-Lys-Pro-Ala-Thr (AKPAT) | 486.56/486.56 | ||
| P11 | 13.92 | MSP13: Tyr-Pro-Ala-Gly-Pro (YPAGP) | 503.55/503.54 |
| P12 | 14.93 | MSP14:Asp-Pro-Thr (DPT) | 331.32/331.32 |
| P13 | 15.02 | MSP15: Phe-Pro-Gly-Pro-Thr (FPGPT) | 517.57/517.57 |
| P14 | 17.18 | MSP16: Gly-Pro-Gly-Pro-Thr (GPGPT) | 427.45/427.45 |
| P15 | 17.99 | MSP17: Gly-Pro-Thr (GPT) | 273.29/273.29 |
| P16 | 20.50 | MSP18: Asp-Pro-Ala-Gly-Pro (DPAGP) | 455.46/455.46 |
| Peptides | EC50 (mg/mL) | |||
|---|---|---|---|---|
| DPPH· | HO· | ABTS+· | ||
| MSP1 | 2.824 ± 0.019 a | 0.150 ± 0.060 a | 0.126 ± 0.0005 a | 3.197 ± 0.036 a |
| MSP2 | 2.993 ± 0.054 a | 0.177 ± 0.035 a,b | 0.112 ± 0.0028 b | 2.337 ± 0.016 b |
| MSP3 | 8.667 ± 0.023 b | 0.475 ± 0.0103 c | 0.166 ± 0.0017 c | 6.693 ± 0.030 c |
| MSP4 | 7.703 ± 0.091 c | 0.244 ± 0.0035 d | 0.397 ± 0.0008 d | 7.613 ± 0.133 d |
| MSP5 | 10.947 ± 0.031 d | 0.652 ± 0.0132 e | 0.239 ± 0.0026 e | 6.641 ± 0.043 c |
| MSP6 | 5.231 ± 0.083 e | 0.349 ± 0.0037 f,g | 0.188 ± 0.0026 f | 5.729 ± 0.025 e,i |
| MSP7 | 1.053 ± 0.003 f | 0.201 ± 0.013 b | 0.245 ± 0.0021 g | 6.188 ± 0.084 f |
| MSP8 | 0.773 ± 0.003 g | 0.183 ± 0.0016 a,b | 0.127 ± 0.0002 a | 4.503 ± 0.040 g |
| MSP9 | 9.420 ± 0.109 h | 0.493 ± 0.0042 c | 0.173 ± 0.0020 h | 8.034 ± 0.124 h |
| MSP10 | 10.417 ± 0.110 i | 0.329 ± 0.004 f,g,h | 0.128 ± 0.0018 a | 6.004 ± 0.087 f,i |
| MSP11 | 8.195 ± 0.271 j | 0.366 ± 0.004 f,g,h | 0.335 ± 0.0027 i | 8.054 ± 0.366 h |
| MSP12 | 13.96 ± 0.284 k | 0.482 ± 0.0309 c | 0.353 ± 0.0006 j | 8.695 ± 0.200 j |
| MSP13 | 2.821 ± 0.012 a | 0.190 ± 0.010 b | 0.107 ± 0.0002 k | 3.839 ± 0.102 k |
| MSP14 | 4.450 ± 0.005 l | 0.353 ± 0.0067 f,g,h | 0.116 ± 0.0002 b | 5.411 ± 0.028 e |
| MSP15 | 11.013 ± 0.042 d | 0.306 ± 0.0025 g | 0.505 ± 0.0058 l | 9.058 ± 0.082 l |
| MSP16 | 13.99 ± 0.046 k | 0.312 ± 0.0065 f,g | 0.208 ± 0.0080 m | 10.89 ± 0.322 m |
| MSP17 | 10.413 ± 0.006 i | 0.439 ± 0.0069 i | 0.185 ± 0.0014 f | 12.387 ± 0.670 n |
| MSP18 | 7.119 ± 0.092 m | 0.332 ± 0.035 f,g,h | 0.166 ± 0.0020 c | 5.621 ± 0.169 e |
| Peptides | Declined Percentage (%) | ||||
|---|---|---|---|---|---|
| pH 3 | pH 5 | pH 7 | pH 9 | pH 11 | |
| MSP1 | −21.77 | −21.10 | 0.00 | −4.97 | −35.26 |
| MSP2 | −36.14 | −33.16 | 0.00 | −2.29 | −73.07 |
| MSP7 | −18.68 | −16.43 | 0.00 | −5.78 | −69.19 |
| MSP8 | −19.97 | −19.47 | 0.00 | −3.85 | −83.05 |
| MSP10 | −28.91 | −24.92 | −5.13 | −7.06 | 0 |
| MSP13 | −26.03 | −21.19 | −0.15 | 0 | −26.03 |
| Declined Percentage (%) | |||
|---|---|---|---|
| Untreated | Treated with Pepsin | Treated with Pepsin and Trypsin | |
| MSP1 | 0.00 | −0.84 | −4.10 |
| MSP2 | 0.00 | −15.13 | −47.59 |
| MSP7 | 0.00 | −24.34 | −49.19 |
| MSP8 | 0.00 | −24.93 | −28.62 |
| MSP10 | 0.00 | −21.98 | −53.33 |
| MSP13 | 0.00 | −28.02 | −35.83 |
| Peptides | Declined Percentage (%) | ||||
|---|---|---|---|---|---|
| 25 °C | 37 °C | 60 °C | 80 °C | 100 °C | |
| MSP1 | 0.00 | −1.48 | −0.82 | −1.15 | −0.82 |
| MSP2 | 0.00 | −1.00 | −1.50 | −3.17 | −3.84 |
| MSP7 | 0.00 | −1.94 | −0.52 | −0.70 | −0.53 |
| MSP8 | 0.00 | 1.02 | 0.34 | −0.17 | −1.70 |
| MSP10 | 0.00 | −0.87 | −1.58 | −2.16 | −2.60 |
| MSP13 | 0.00 | 0.00 | 0.00 | 0.61 | −0.46 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sheng, Y.; Wang, W.-Y.; Wu, M.-F.; Wang, Y.-M.; Zhu, W.-Y.; Chi, C.-F.; Wang, B. Eighteen Novel Bioactive Peptides from Monkfish (Lophius litulon) Swim Bladders: Production, Identification, Antioxidant Activity, and Stability. Mar. Drugs 2023, 21, 169. https://doi.org/10.3390/md21030169
Sheng Y, Wang W-Y, Wu M-F, Wang Y-M, Zhu W-Y, Chi C-F, Wang B. Eighteen Novel Bioactive Peptides from Monkfish (Lophius litulon) Swim Bladders: Production, Identification, Antioxidant Activity, and Stability. Marine Drugs. 2023; 21(3):169. https://doi.org/10.3390/md21030169
Chicago/Turabian StyleSheng, Yan, Wan-Yi Wang, Ming-Feng Wu, Yu-Mei Wang, Wang-Yu Zhu, Chang-Feng Chi, and Bin Wang. 2023. "Eighteen Novel Bioactive Peptides from Monkfish (Lophius litulon) Swim Bladders: Production, Identification, Antioxidant Activity, and Stability" Marine Drugs 21, no. 3: 169. https://doi.org/10.3390/md21030169
APA StyleSheng, Y., Wang, W. -Y., Wu, M. -F., Wang, Y. -M., Zhu, W. -Y., Chi, C. -F., & Wang, B. (2023). Eighteen Novel Bioactive Peptides from Monkfish (Lophius litulon) Swim Bladders: Production, Identification, Antioxidant Activity, and Stability. Marine Drugs, 21(3), 169. https://doi.org/10.3390/md21030169