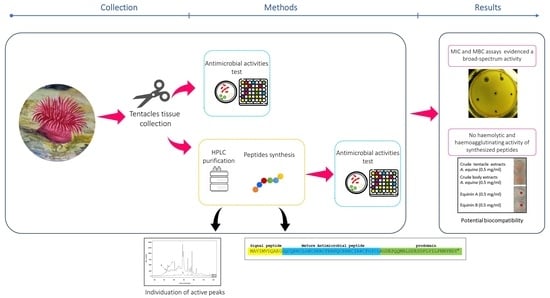
Equinins as Novel Broad-Spectrum Antimicrobial Peptides Isolated from the Cnidarian Actinia equina (Linnaeus, 1758)
Abstract
:1. Introduction
2. Results
2.1. Antimicrobial Activity of A. equina Extracts
2.2. Fractionation of A. equina Tentacle Acid Extracts
2.3. Reversed-Phase Chromatography
2.4. Sequencing
2.5. Antibacterial Activity of Newly Synthesized Peptides: Equinin A and Equinin B
2.6. Haemolytic Activity
2.7. AMP Predictions through In Silico Analysis
3. Discussion
4. Materials and Methods
4.1. A. equina Collection and Extract Preparation
4.2. Microbial Strains
4.3. Evaluation of Antibacterial Activity
4.4. Determination of MICs and MBC
4.5. Solid Phase Extraction and Reversed-Phase HPLC Purification
4.6. Amino Acid Sequencing
4.7. Peptide Synthesis
4.8. Haemolytic Activity
4.9. In Silico Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Fischer, W.; Bauchot, M.L.; Schneider, M. Fiches FAO d’ Identification Des Espèces Pour Les Besoins de La Pêche. (Revision 1). Méditerranée et Mer Noire. Zone de Pêche 37. Volume I. Végëtaux et Invertébrés; Publication Préparée Par La FAO, Résultant d’un Accord Entre La FAO et La Commission Des C; FAO: Rome, Italy, 1987; Volume 760. [Google Scholar]
- Steiner, H.; Hultmark, D.; Engström, Å.; Bennich, H.; Boman, H.G. Sequence and Specificity of Two Antibacterial Proteins Involved in Insect Immunity. Nature 1981, 292, 246–248. [Google Scholar] [CrossRef] [PubMed]
- Boman, H.G. Antibacterial Peptides: Basic Facts and Emerging Concepts. J. Intern. Med. 2003, 254, 197–215. [Google Scholar] [CrossRef] [PubMed]
- Bulet, P.; Stöcklin, R.; Menin, L. Anti-microbial Peptides: From Invertebrates to Vertebrates. Immunol. Rev. 2004, 198, 169–184. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Gao, J.; Tong, P.; Chen, H. Antimicrobial Peptides: Action Mechanism, Application and Improvement Strategy. Chin. J. Anim. Nutr. 2017, 29, 3885–3892. [Google Scholar]
- Wu, R.; Patocka, J.; Nepovimova, E.; Oleksak, P.; Valis, M.; Wu, W.; Kuca, K. Marine Invertebrate Peptides: Antimicrobial Peptides. Front. Microbiol. 2021, 12, 785085. [Google Scholar] [CrossRef] [PubMed]
- Bartlett, T.C.; Cuthbertson, B.J.; Shepard, E.F.; Chapman, R.W.; Gross, P.S.; Warr, G.W. Crustins, Homologues of an 11.5-KDa Antibacterial Peptide, from Two Species of Penaeid Shrimp, Litopenaeus vannamei and Litopenaeus setiferus. Mar. Biotechnol. 2002, 4, 278–293. [Google Scholar] [CrossRef]
- Chisholm, J.R.S.; Smith, V.J. Antibacterial Activity in the Haemocytes of the Shore Crab, Carcinus maenas. J. Mar. Biol. Assoc. 1992, 72, 529–542. [Google Scholar] [CrossRef]
- Ovchinnikova, T.V.; Balandin, S.V.; Aleshina, G.M.; Tagaev, A.A.; Leonova, Y.F.; Krasnodembsky, E.D.; Men’shenin, A.V.; Kokryakov, V.N. Aurelin, a Novel Antimicrobial Peptide from Jellyfish Aurelia aurita with Structural Features of Defensins and Channel-Blocking Toxins. Biochem. Biophys. Res. Commun. 2006, 348, 514–523. [Google Scholar] [CrossRef] [PubMed]
- Bosch, T.C.G.; Augustin, R.; Anton-Erxleben, F.; Fraune, S.; Hemmrich, G.; Zill, H.; Rosenstiel, P.; Jacobs, G.; Schreiber, S.; Leippe, M.; et al. Uncovering the Evolutionary History of Innate Immunity: The Simple Metazoan Hydra Uses Epithelial Cells for Host Defence. Dev. Comp. Immunol. 2009, 33, 559–569. [Google Scholar] [CrossRef]
- Tejuca, M.; Dalla Serra, M.; Ferreras, M.; Lanio, M.E.; Menestrina, G. Mechanism of Membrane Permeabilization by Sticholysin I, a Cytolysin Isolated from the Venom of the Sea Anemone Stichodactyla helianthus. Biochemistry 1996, 35, 14947–14957. [Google Scholar] [CrossRef]
- Trapani, M.R.; Parisi, M.G.; Toubiana, M.; Coquet, L.; Jouenne, T.; Roch, P.; Cammarata, M. First Evidence of Antimicrobial Activity of Neurotoxin 2 from Anemonia sulcata (Cnidaria). Invertebr. Surviv. J. 2014, 11, 182–191. [Google Scholar]
- Nicosia, A.; Mikov, A.; Cammarata, M.; Colombo, P.; Andreev, Y.; Kozlov, S.; Cuttitta, A. The Anemonia viridis Venom: Coupling Biochemical Purification and Rna-Seq for Translational Research. Mar. Drugs 2018, 16, 407. [Google Scholar] [CrossRef] [PubMed]
- Tincu, J.A.; Taylor, S.W. Antimicrobial Peptides from Marine Invertebrates. Antimicrob. Agents Chemother. 2004, 48, 3645–3654. [Google Scholar] [CrossRef] [PubMed]
- Boman, H.G. Gene-Encoded Peptide Antibiotics and the Concept of Innate Immunity: An Update Review. Scand. J. Immunol. 1998, 48, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Lehrer, R.I.; Ganz, T. Antimicrobial Peptides in Mammalian and Insect Host Defence. Curr. Opin. Immunol. 1999, 11, 23–27. [Google Scholar] [CrossRef] [PubMed]
- Hancock, R.E.W.; Scott, M.G. The Role of Antimicrobial Peptides in Animal Defenses. Proc. Natl. Acad. Sci. USA 2000, 97, 8856–8861. [Google Scholar] [CrossRef] [PubMed]
- van der Does, A.M.; Hiemstra, P.S.; Mookherjee, N. Antimicrobial Host Defence Peptides: Immunomodulatory Functions and Translational Prospects. In Antimicrobial Peptides: Basics for Clinical Application; Springer: Berlin/Heidelberg, Germany, 2019; pp. 149–171. [Google Scholar]
- Kumar, P.; Kizhakkedathu, J.N.; Straus, S.K. Antimicrobial Peptides: Diversity, Mechanism of Action and Strategies to Improve the Activity and Biocompatibility in Vivo. Biomolecules 2018, 8, 4. [Google Scholar] [CrossRef] [PubMed]
- Smith, V.J.; Desbois, A.P.; Dyrynda, E.A. Conventional and Unconventional Antimicrobials from Fish, Marine Invertebrates and Micro-Algae. Mar. Drugs 2010, 8, 1213–1262. [Google Scholar] [CrossRef] [PubMed]
- Punginelli, D.; Catania, V.; Vazzana, M.; Mauro, M.; Spinello, A.; Barone, G.; Barberi, G.; Fiorica, C.; Vitale, M.; Cunsolo, V.; et al. A Novel Peptide with Antifungal Activity from Red Swamp Crayfish Procambarus clarkii. Antibiotics 2022, 11, 1792. [Google Scholar] [CrossRef]
- Li, C.; Sutherland, D.; Hammond, S.A.; Yang, C.; Taho, F.; Bergman, L.; Houston, S.; Warren, R.L.; Wong, T.; Hoang, L.M.N.; et al. AMPlify: Attentive Deep Learning Model for Discovery of Novel Antimicrobial Peptides Effective against WHO Priority Pathogens. BMC Genom. 2022, 23, 77. [Google Scholar] [CrossRef]
- Trapani, M.R.; Parisi, M.G.; Maisano, M.; Mauceri, A.; Cammarata, M. Old Weapons for New Wars: Bioactive Molecules From Cnidarian Internal Defense Systems. Cent. Nerv. Syst. Agents Med. Chem. 2016, 16, 183–196. [Google Scholar] [CrossRef]
- Lassen, S.; Helmholz, H.; Ruhnau, C.; Prange, A. A Novel Proteinaceous Cytotoxin from the Northern Scyphozoa Cyanea capillata (L.) with Structural Homology to Cubozoan Haemolysins. Toxicon 2011, 57, 721–729. [Google Scholar] [CrossRef] [PubMed]
- Morabito, R.; Condello, S.; Currò, M.; Marino, A.; Ientile, R.; La Spada, G. Oxidative Stress Induced by Crude Venom from the Jellyfish Pelagia noctiluca in Neuronal-like Differentiated SH-SY5Y Cells. Toxicol. Vitr. 2012, 26, 694–699. [Google Scholar] [CrossRef] [PubMed]
- Destoumieux-Garzón, D.; Rosa, R.D.; Schmitt, P.; Barreto, C.; Vidal-Dupiol, J.; Mitta, G.; Gueguen, Y.; Bachère, E. Antimicrobial Peptides in Marine Invertebrate Health and Disease. Philos. Trans. R. Soc. B Biol. Sci. 2016, 371, 20150300. [Google Scholar] [CrossRef] [PubMed]
- Torri, L.; Tuccillo, F.; Bonelli, S.; Piraino, S.; Leone, A. The Attitudes of Italian Consumers towards Jellyfish as Novel Food. Food Qual. Prefer. 2020, 79, 103782. [Google Scholar] [CrossRef]
- De Zoysa, M. Medicinal Benefits of Marine Invertebrates: Sources for Discovering Natural Drug Candidates. Adv. Food Nutr. Res. 2012, 65, 153–169. [Google Scholar] [CrossRef] [PubMed]
- Youssef, J.; Keller, S.; Spence, C. Making Sustainable Foods (Such as Jellyfish) Delicious. Int. J. Gastron. Food Sci. 2019, 16, 100141. [Google Scholar] [CrossRef]
- Kalina, R.S.; Peigneur, S.; Zelepuga, E.A.; Dmitrenok, P.S.; Kvetkina, A.N.; Kim, N.Y.; Leychenko, E.V.; Tytgat, J.; Kozlovskaya, E.P.; Monastyrnaya, M.M. New Insights into the Type II Toxins from the Sea Anemone Heteractis crispa. Toxins 2020, 12, 44. [Google Scholar] [CrossRef]
- Liao, Q.; Feng, Y.; Yang, B.; Lee, S.M.-Y. Cnidarian Peptide Neurotoxins: A New Source of Various Ion Channel Modulators or Blockers against Central Nervous Systems Disease. Drug Discov. Today 2019, 24, 189–197. [Google Scholar] [CrossRef]
- Augustin, R.; Anton-Erxleben, F.; Jungnickel, S.; Hemmrich, G.; Spudy, B.; Podschun, R.; Bosch, T.C.G. Activity of the Novel Peptide Arminin against Multiresistant Human Pathogens Shows the Considerable Potential of Phylogenetically Ancient Organisms as Drug Sources. Antimicrob. Agents Chemother. 2009, 53, 5245–5250. [Google Scholar] [CrossRef]
- Liang, X.; Wang, R.; Dou, W.; Zhao, L.; Zhou, L.; Zhu, J.; Wang, K.; Yan, J. Arminin 1a-C, a Novel Antimicrobial Peptide from Ancient Metazoan Hydra, Shows Potent Antileukemia Activity against Drug-Sensitive and Drug-Resistant Leukemia Cells. Drug Des. Devel. Ther. 2018, 12, 3691–3703. [Google Scholar] [CrossRef] [PubMed]
- Kelman, D.; Kashman, Y.; Rosenberg, E.; Kushmaro, A.; Loya, Y. Antimicrobial Activity of Red Sea Corals. Mar. Biol. 2006, 149, 357–363. [Google Scholar] [CrossRef]
- Lei, J.; Sun, L.C.; Huang, S.; Zhu, C.; Li, P.; He, J.; Mackey, V.; Coy, D.H.; He, Q.Y. The Antimicrobial Peptides and Their Potential Clinical Applications. Am. J. Transl. Res. 2019, 11, 3919–3931. [Google Scholar] [PubMed]
- Palla, F.; Barresi, G.; Giordano, A.; Schiavone, S.; Trapani, M.R.; Rotolo, V.; Parisi, M.G. Cold-Active Molecules for a Sustainable Preservation and Restoration of Historic-Artistic Manufacts. Int. J. Conserv. Sci. 2016, 7, 239–246. [Google Scholar]
- Barresi, G.; Di Carlo, E.; Trapani, M.R.; Parisi, M.G.; Chille, C.; Mule, M.F.; Cammarata, M.; Palla, F. Marine Organisms as Source of Bioactive Molecules Applied in Restoration Projects. Herit. Sci. 2015, 3, 17. [Google Scholar] [CrossRef]
- Stabili, L.; Schirosi, R.; Parisi, M.G.; Piraino, S.; Cammarata, M. The Mucus of Actinia equina (Anthozoa, Cnidaria): An Unexplored Resource for Potential Applicative Purposes. Mar. Drugs 2015, 13, 5276–5296. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Stabili, L.; Licciano, M.; Giangrande, A.; Gerardi, C.; De Pascali, S.A.; Fanizzi, F.P. First Insight on the Mucus of the Annelid Myxicola infundibulum (Polychaeta, Sabellidae) as a Potential Prospect for Drug Discovery. Mar. Drugs 2019, 17, 396. [Google Scholar] [CrossRef] [PubMed]
- Balouiri, M.; Sadiki, M.; Ibnsouda, S.K. Methods for in Vitro Evaluating Antimicrobial Activity: A Review. J. Pharm. Anal. 2016, 6, 71–79. [Google Scholar] [CrossRef]
- Ballarin, L.; Cammarata, M.; Cima, F.; Grimaldi, A.; Lorenzon, S.; Malagoli, D.; Ottaviani, E. Immune-Neuroendocrine Biology of Invertebrates: A Collection of Methods. Invertebr. Surviv. J. 2008, 5, 192–215. [Google Scholar]
- Wang, Z.; Wang, G. APD: The Antimicrobial Peptide Database. Nucleic Acids Res. 2004, 32, D590–D592. [Google Scholar] [CrossRef] [PubMed]
- Thomas, S.; Karnik, S.; Barai, R.S.; Jayaraman, V.K.; Idicula-Thomas, S. CAMP: A Useful Resource for Research on Antimicrobial Peptides. Nucleic Acids Res. 2010, 38, D774–D780. [Google Scholar] [CrossRef] [PubMed]
- Torrent, M.; Nogués, V.M.; Boix, E. A Theoretical Approach to Spot Active Regions in Antimicrobial Proteins. BMC Bioinform. 2009, 10, 373. [Google Scholar] [CrossRef] [PubMed]
- Torrent, M.; Di Tommaso, P.; Pulido, D.; Nogués, M.V.; Notredame, C.; Boix, E.; Andreu, D. AMPA: An Automated Web Server for Prediction of Protein Antimicrobial Regions. Bioinformatics 2012, 28, 130–131. [Google Scholar] [CrossRef] [PubMed]
- White, S.H.; Wimley, W.C. Membrane Protein Folding and Stability: Physical Principles. Annu. Rev. Biophys. Biomol. Struct. 1999, 28, 319–365. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology Modelling of Protein Structures and Complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Studer, G.; Rempfer, C.; Waterhouse, A.M.; Gumienny, R.; Haas, J.; Schwede, T. QMEANDisCo-Distance Constraints Applied on Model Quality Estimation. Bioinformatics 2020, 36, 1765–1771. [Google Scholar] [CrossRef]








| Minimum Inhibitory Concentration (MIC) in mg/mL | ||
|---|---|---|
| Tentacle Acid Extracts | Body Acid Extracts | |
| E. coli ATCC 25922 | 0.25 | 0.5 |
| M. lysodeikticus ATCC 4698 | 0.125 | 0.5 |
| V. alginolyticus ATCC 17749 | 0.5 | 0.5 |
| P. aeruginosa ATCC 15442 | 0.25 | - |
| Tentacle Extracts | Peptide | Identified Sequences | Percentage Similarity with Already Described AMPs | Activity of AMPs with Better Similarity | References |
| Equinin A | AVDKGGGKAEKKDGNRKKKLAGGEGGG | 42.42% RaCa-1, Palustrin (frogs, amphibians) | Anti-Gram+ & Gram- | [22] | |
| Equinin B | GQCQRKCLGHCSKKCPKHPQCRKRCIRRCFGYCL | 37.78% Aurelin (Jellyfish, Cnidaria) | Anti-Gram+ & Gram- | [9] |
| Equinin A | Equinin B | |
|---|---|---|
| Peptide residues | 27 | 34 |
| Monoisotopic Theoretical mass (Da) | 2612.91 | 3934.827 |
| Net charge | +4 | +11.5 |
| Wimley-White whole residues (kcal/mol) | 14.77 | 8.97 |
| Hydrophobic ratio (%) | 19 | 32 |
| Protein-binding potential Boman index (kcal/mol) | 2.54 | 3.24 |
| GRAVY (i.e., the grand average hydropathy value of the peptide) | −1.46 | −1.11 |
| Minimum Inhibitory Concentration (MIC) in mg/mL | ||
|---|---|---|
| Equinin A | Equinin B | |
| E. coli ATCC 25922 | >1 | 0.25 |
| M. lysodeikticus ATCC 4698 | >1 | 0.25 |
| V. alginolyticus ATTC 17749 | >1 | 0.25 |
| Properties | Equinin A | Equinin B |
|---|---|---|
| CPP (cell penetrating peptides) | Not CPP | Not CPP |
| Antibacterial activity | 0.91 | 1 |
| Antifungal activity | 0.8 | 0.99 |
| Antiviral activity | 0.37 | 0.96 |
| Antibiofilm activity | inactive | active |
| Degradation by trypsin | Yes | Yes |
| Degradation by pepsin (pH = 1.3) | No | Yes |
| Degradation by pepsin (pH > 2) | No | Yes |
| Extracts (mg/mL) | Fractions | ||
|---|---|---|---|
| 10% | 40% | 60% | |
| Tentacles | 0.98 | 7.83 | 0.2 |
| Body | 1.29 | 3.49 | 0.13 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
La Corte, C.; Catania, V.; Dara, M.; Parrinello, D.; Staropoli, M.; Trapani, M.R.; Cammarata, M.; Parisi, M.G. Equinins as Novel Broad-Spectrum Antimicrobial Peptides Isolated from the Cnidarian Actinia equina (Linnaeus, 1758). Mar. Drugs 2024, 22, 172. https://doi.org/10.3390/md22040172
La Corte C, Catania V, Dara M, Parrinello D, Staropoli M, Trapani MR, Cammarata M, Parisi MG. Equinins as Novel Broad-Spectrum Antimicrobial Peptides Isolated from the Cnidarian Actinia equina (Linnaeus, 1758). Marine Drugs. 2024; 22(4):172. https://doi.org/10.3390/md22040172
Chicago/Turabian StyleLa Corte, Claudia, Valentina Catania, Mariano Dara, Daniela Parrinello, Mariele Staropoli, Maria Rosa Trapani, Matteo Cammarata, and Maria Giovanna Parisi. 2024. "Equinins as Novel Broad-Spectrum Antimicrobial Peptides Isolated from the Cnidarian Actinia equina (Linnaeus, 1758)" Marine Drugs 22, no. 4: 172. https://doi.org/10.3390/md22040172
APA StyleLa Corte, C., Catania, V., Dara, M., Parrinello, D., Staropoli, M., Trapani, M. R., Cammarata, M., & Parisi, M. G. (2024). Equinins as Novel Broad-Spectrum Antimicrobial Peptides Isolated from the Cnidarian Actinia equina (Linnaeus, 1758). Marine Drugs, 22(4), 172. https://doi.org/10.3390/md22040172