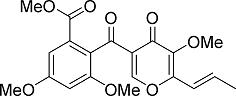
3-O-Methylfunicone, a Selective Inhibitor of Mammalian Y-Family DNA Polymerases from an Australian Sea Salt Fungal Strain
Abstract
:1. Introduction
2. Results and Discussion
2.1. Purification of a DNA Polymerase Inhibitor, 3-O-Methylfunicone (Compound 1), from a Marine Fungal Strain
2.2. Effect of Isolated Compound 1 on the Activities of DNA Polymerases and Other DNA Metabolic Enzymes
2.3. Mode of DNA Pol κ Inhibition by Compound 1
2.4. Inhibitory Effect of Isolated Compound 1 on Cultured Human Cancer Cells
3. Conclusions
4. Experimental
4.1. Isolation and Cultivation of Marine Fungal Strains from Australian Sea Salt
4.2. Extraction and Purification of Compound from Marine Fungal Strains
4.3. Structure Determination of Isolated Compound
4.4. Materials
4.5. Enzymes
4.6. DNA Polymerase Assays
4.7. Other DNA Metabolic Enzymes Assays
4.8. Investigation of Growth on Cultured Human Cancer Cells
4.9. UV-Treated Clonogenic Assay
Acknowledgments
- Samples Availability: Available from the corresponding author.
References and Notes
- DePamphilis, ML. DNA Replication in Eukaryotic Cells; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1996. [Google Scholar]
- Seto, H; Hatanaka, H; Kimura, S; Oshige, M; Tsuya, Y; Mizushina, Y; Sawado, T; Aoyagi, N; Matsumoto, T; Hashimoto, J; Sakaguchi, K. Purification and characterization of a 100 kDa DNA polymerase from cauliflower inflorescence. Biochem J 1998, 332, 557–563. [Google Scholar]
- Hubscher, U; Maga, G; Spadari, S. Eukaryotic DNA polymerases. Ann Rev Biochem 2002, 71, 133–163. [Google Scholar]
- Bebenek, K; Kunkel, TA. DNA repair and replication. In Advances in Protein Chemistry; Yang, W, Ed.; Elsevier: San Diego, CA, USA, 2004; Volume 69, pp. 137–165. [Google Scholar]
- Takata, K; Shimizu, T; Iwai, S; Wood, RD. Human DNA polymerase N (POLN) is a low fidelity enzyme capable of error-free bypass of 5S-thymine glycol. J Biol Chem 2006, 281, 23445–23455. [Google Scholar]
- Friedberg, EC; Feaver, WJ; Gerlach, VL. The many faces of DNA polymerases: Strategies for mutagenesis and for mutational avoidance. Proc Natl Acad Sci USA 2000, 97, 5681–5683. [Google Scholar]
- Mizushina, Y; Tanaka, N; Yagi, H; Kurosawa, T; Onoue, M; Seto, H; Horie, T; Aoyagi, N; Yamaoka, M; Matsukage, A; Yoshida, S; Sakaguchi, K. Fatty acids selectively inhibit eukaryotic DNA polymerase activities in vitro. Biochim Biophys Acta 1996, 1308, 256–262. [Google Scholar]
- Mizushina, Y; Yoshida, S; Matsukage, A; Sakaguchi, K. The inhibitory action of fatty acids on DNA polymerase β. Biochim Biophys Acta 1997, 1336, 509–521. [Google Scholar]
- Mizushina, Y; Takahashi, N; Ogawa, A; Tsurugaya, K; Koshino, H; Takemura, M; Yoshida, S; Matsukage, A; Sugawara, F; Sakaguchi, K. The cyanogenic glucoside, prunasin (D-mandelonitrile-β-D-glucoside), is a novel inhibitor of DNA polymerase β. J Biochem (Tokyo) 1999, 126, 430–436. [Google Scholar]
- Mizushina, Y; Ohkubo, T; Sugawara, F; Sakaguchi, K. Structure of lithocholic acid binding to the N-terminal 8-kDa domain of DNA polymerase β. Biochemistry 2000, 39, 12606–12613. [Google Scholar]
- Mizushina, Y; Kamisuki, S; Mizuno, T; Takemura, M; Asahara, H; Linn, S; Yamaguchi, T; Matsukage, A; Hanaoka, F; Yoshida, S; Saneyoshi, M; Sugawara, F; Sakaguchi, K. Dehydroaltenusin, a mammalian DNA polymerase α inhibitor. J Biol Chem 2000, 275, 33957–33961. [Google Scholar]
- Mizushina, Y; Kamisuki, S; Kasai, N; Shimazaki, N; Takemura, M; Asahara, H; Linn, S; Yoshida, S; Matsukage, A; Koiwai, O; Sugawara, F; Yoshida, H; Sakaguchi, K. A plant phytotoxin, solanapyrone A, is an inhibitor of DNA polymerase β and λ. J Biol Chem 2002, 277, 630–638. [Google Scholar]
- Mizushina, Y; Xu, X; Asahara, H; Takeuchi, R; Oshige, M; Shimazaki, N; Takemura, M; Yamaguchi, T; Kuroda, K; Linn, S; Yoshida, H; Koiwai, O; Saneyoshi, M; Sugawara, F; Sakaguchi, K. A sulphoquinovosyl diacylglycerol is a DNA polymerase ɛ inhibitor. Biochem J 2003, 370, 299–305. [Google Scholar]
- Kuriyama, I; Asano, N; Kato, I; Ikeda, K; Takemura, M; Yoshida, H; Sakaguchi, K; Mizushina, Y. Dipeptide alcohol-based inhibitors of eukaryotic DNA polymerase α. Bioorg Med Chem 2005, 13, 2187–2196. [Google Scholar]
- Kamisuki, S; Ishimaru, C; Onoda, K; Kuriyama, I; Ida, N; Sugawara, F; Yoshida, H; Mizushina, Y. Nodulisporol and Nodulisporone, novel specific inhibitors of human DNA polymerase λ from a fungus, Nodulisporium sp. Bioorg Med Chem 2007, 15, 3109–3114. [Google Scholar]
- Naganuma, M; Nishida, M; Kuramochi, K; Sugawara, F; Yoshida, H; Mizushina, Y. 1-Deoxyrubralactone, a novel specific inhibitor of families X and Y of eukaryotic DNA polymerases from a fungal strain derived from sea algae. Bioorg Med Chem 2008, 16, 2939–2944. [Google Scholar]
- Kimura, T; Nishida, M; Kuramochi, K; Sugawara, F; Yoshida, H; Mizushina, Y. Novel azaphilones, kasanosins A and B, which are specific inhibitors of eukaryotic DNA polymerases β and λ from Talaromyces sp. Bioorg Med Chem 2008, 16, 4594–4599. [Google Scholar]
- Nishida, M; Ida, N; Horio, M; Takeuchi, T; Kamisuki, S; Murata, H; Kuramochi, K; Sugawara, F; Yoshida, H; Mizushina, Y. Hymenoic acid, a novel specific inhibitor of human DNA polymerase λ from a fungus of Hymenochaetaceae sp. Bioorg Med Chem 2008, 16, 5115–5122. [Google Scholar]
- Kimura, T; Takeuchi, T; Kumamoto-Yonezawa, Y; Ohashi, E; Ohmori, H; Masutani, C; Hanaoka, F; Sugawara, F; Yoshida, H; Mizushina, Y. Penicilliols A and B, novel inhibitors specific to mammalian Y-family DNA polymerases. Bioorg Med Chem 2009, 17, 1811–1816. [Google Scholar]
- Oguro, M; Suzuki-Hori, C; Nagano, H; Mano, Y; Ikegami, S. The mode of inhibitory action by aphidicolin on eukaryotic DNA polymerase α. Eur J Biochem 1979, 97, 603–607. [Google Scholar]
- Prakash, S; Johnson, RE; Prakash, L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu Rev Biochem 2005, 74, 317–353. [Google Scholar]
- Chen, YW; Cleaver, JE; Hanaoka, F; Chang, CF; Chou, KM. A novel role of DNA polymerase eta in modulating cellular sensitivity to chemotherapeutic agents. Mol Cancer Res 2006, 4, 257–265. [Google Scholar]
- Stefano, SD; Nicoletti, R; Milone, A; Zambardino, S. 3-o-Methylfunicone, a fungitoxic metabolite produced by the fungus. Penicillium pinophilum Phytochemistry 1999, 52, 1399–1401. [Google Scholar]
- Tamai, K; Kojima, K; Hanaichi, T; Masaki, S; Suzuki, M; Umekawa, H; Yoshida, S. Structural study of immunoaffinity-purified DNA polymerase α-DNA primase complex from calf thymus. Biochim Biophys Acta 1988, 950, 263–273. [Google Scholar]
- Date, T; Yamaguchi, M; Hirose, F; Nishimoto, Y; Tanihara, K; Matsukage, A. Expression of active rat DNA polymerase β in Escherichia coli. Biochemistry 1988, 27, 2983–2990. [Google Scholar]
- Umeda, S; Muta, T; Ohsato, T; Takamatsu, C; Hamasaki, N; Kang, D. The D-loop structure of human mtDNA is destabilized directly by 1-methyl-4-phenylpyridinium ion (MPP+), a parkinsonism-causing toxin. Eur J Biochem 2000, 267, 200–206. [Google Scholar]
- Oshige, M; Takeuchi, R; Ruike, R; Kuroda, K; Sakaguchi, K. Subunit protein-affinity isolation of Drosophila DNA polymerase catalytic subunit. Protein Expr Purif 2004, 35, 248–256. [Google Scholar]
- Kusumoto, R; Masutani, C; Shimmyo, S; Iwai, S; Hanaoka, F. DNA binding properties of human DNA polymerase η: implications for fidelity and polymerase switching of translesion synthesis. Genes Cells 2004, 9, 1139–1150. [Google Scholar]
- Ohashi, E; Murakumo, Y; Kanjo, N; Akagi, J; Masutani, C; Hanaoka, F; Ohmori, H. Interaction of hREV1 with three human Y-family DNA polymerases. Genes Cells 2004, 9, 523–531. [Google Scholar]
- Shimazaki, N; Yoshida, K; Kobayashi, T; Toji, S; Tamai, T; Koiwai, O. Over-expression of human DNA polymerase λ in E. coli and characterization of the recombinant enzyme. Genes Cells 2000, 7, 639–651. [Google Scholar]
- Yamaguchi, T; Saneyoshi, M; Takahashi, H; Hirokawa, S; Amano, R; Liu, X; Inomata, M; Maruyama, T. Synthetic Nucleoside and Nucleotides. 43. Inhibition of vertebrate telomerases by carbocyclic oxetanocin G (C.OXT-G) triphosphate analogues and influence of C.OXT-G treatment on telomere length in human HL60 cells. Nucleos Nucleot Nucleic Acids 2006, 25, 539–551. [Google Scholar]
- Aoyagi, N; Matsuoka, S; Furunobu, A; Matsukage, A; Sakaguchi, K. Drosophila DNA polymerase δ. Purification and characterization. J Biol Chem 1994, 269, 6045–6050. [Google Scholar]
- Aoyagi, N; Oshige, M; Hirose, F; Kuroda, K; Matsukage, A; Sakaguchi, K. DNA polymerase ɛ from Drosophila melanogaster. Biochem Biophys Res Commun 1997, 230, 297–301. [Google Scholar]
- Sakaguchi, K; Hotta, Y; Stern, H. Chromatin-associated DNA polymerase activity in meiotic cells of lily and mouse. Cell Struct Funct 1980, 5, 323–334. [Google Scholar]
- Uchiyama, Y; Kimura, S; Yamamoto, T; Ishibashi, T; Sakaguchi, K. Plant DNA polymerase λ, a DNA repair enzyme that functions in plant meristematic and meiotic tissues. Eur J Biochem 2004, 271, 2799–2807. [Google Scholar]
- Ogawa, A; Murate, T; Suzuki, M; Nimura, Y; Yoshida, S. Lithocholic acid, a putative tumor promoter, inhibits mammalian DNA polymerase β. Jpn J Cancer Res 1998, 89, 1154–1159. [Google Scholar]
- Tamiya-Koizumi, K; Murate, T; Suzuki, M; Simbulan, CG; Nakagawa, M; Takamura, M; Furuta, K; Izuta, S; Yoshida, S. Inhibition of DNA primase by sphingosine and its analogues parallels with their growth suppression of cultured human leukemic cells. Biochem Mol Biol Int 1997, 41, 1179–1189. [Google Scholar]
- Oda, M; Ueno, T; Kasai, N; Takahashi, H; Yoshida, H; Sugawara, F; Sakaguchi, K; Hayashi, H; Mizushina, Y. Inhibition of telomerase by linear-chain fatty acids: A structural analysis. Biochem J 2002, 367, 329–334. [Google Scholar]
- Nakayama, C; Saneyoshi, M. Inhibitory effects of 9-β-D-xylofuranosyladenine 5′-triphosphate on DNA-dependent RNA polymerase I and II from cherry salmon (Oncorhynchus masou). J Biochem (Tokyo) 1985, 97, 1385–1389. [Google Scholar]
- Mizushina, Y; Dairaku, I; Yanaka, N; Takeuchi, T; Ishimaru, C; Sugawara, F; Yoshida, H; Kato, N. Inhibitory action of polyunsaturated fatty acids on IMP dehydrogenase. Biochimie 2007, 89, 581–590. [Google Scholar]
- Ishimaru, C; Yonezawa, Y; Kuriyama, I; Nishida, M; Yoshida, H; Mizushina, Y. Inhibitory effects of cholesterol derivatives on DNA polymerase and topoisomerase activities, and human cancer cell growth. Lipids 2008, 43, 373–382. [Google Scholar]
- Soltis, DA; Uhlenbeck, OC. Isolation and characterization of two mutant forms of T4 polynucleotide kinase. J Biol Chem 1982, 257, 11332–11339. [Google Scholar]
- Lu, BC; Sakaguchi, K. An endo-exonuclease from meiotic tissues of the basidiomycete Coprinus cinereus: Its purification and characterization. J Biol Chem 1991, 266, 21060–21066. [Google Scholar]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods 1983, 65, 55–63. [Google Scholar]








© 2009 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Mizushina, Y.; Motoshima, H.; Yamaguchi, Y.; Takeuchi, T.; Hirano, K.; Sugawara, F.; Yoshida, H. 3-O-Methylfunicone, a Selective Inhibitor of Mammalian Y-Family DNA Polymerases from an Australian Sea Salt Fungal Strain. Mar. Drugs 2009, 7, 624-639. https://doi.org/10.3390/md7040624
Mizushina Y, Motoshima H, Yamaguchi Y, Takeuchi T, Hirano K, Sugawara F, Yoshida H. 3-O-Methylfunicone, a Selective Inhibitor of Mammalian Y-Family DNA Polymerases from an Australian Sea Salt Fungal Strain. Marine Drugs. 2009; 7(4):624-639. https://doi.org/10.3390/md7040624
Chicago/Turabian StyleMizushina, Yoshiyuki, Hirohisa Motoshima, Yasuhiro Yamaguchi, Toshifumi Takeuchi, Ken Hirano, Fumio Sugawara, and Hiromi Yoshida. 2009. "3-O-Methylfunicone, a Selective Inhibitor of Mammalian Y-Family DNA Polymerases from an Australian Sea Salt Fungal Strain" Marine Drugs 7, no. 4: 624-639. https://doi.org/10.3390/md7040624
APA StyleMizushina, Y., Motoshima, H., Yamaguchi, Y., Takeuchi, T., Hirano, K., Sugawara, F., & Yoshida, H. (2009). 3-O-Methylfunicone, a Selective Inhibitor of Mammalian Y-Family DNA Polymerases from an Australian Sea Salt Fungal Strain. Marine Drugs, 7(4), 624-639. https://doi.org/10.3390/md7040624