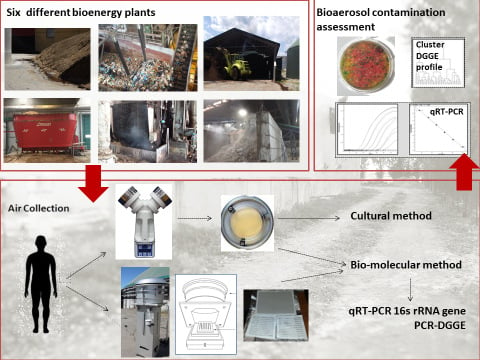
Monitoring of Air Microbial Contaminations in Different Bioenergy Facilities Using Cultural and Biomolecular Methods
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Descriptions
Sampling Points
2.2. Sampling Methods
2.2.1. Particulate Matter (PM) < 0.49 Extraction
2.2.2. Bioaerosol Sampling
2.3. DNA Extraction and Real-Time PCR
- For filter extraction (24 total samples) 1/16 of glass microfiber filter (corresponding to 19 m3 of sampled air) was used. Filters are made of porous material and in the first step of extraction, more lyses buffer was used (2 mL);
- For LMA extraction (32 total samples) the agarized medium was depolymerized by a microwave for a few seconds; the extraction was performed starting from 200 µL, corresponding to 0.1 m3 of sampled air.
2.4. PCR-DGGE and Sequencing
2.5. Data Analysis and Statistics
3. Results and Discussion
3.1. Cultural Method vs. Biomolecular Method
3.2. Biodiversity Analysis
3.3. Sequencing Analysis
3.4. Strong Point and Limitations
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- ISPRA. Rapporto Rifiuti Urbani - Edizione 2018; ISPRA: Rome, Italy, 2018.
- Douglas, P.; Hayes, E.T.; Williams, W.B.; Tyrrel, S.F.; Kinnersley, R.P.; Walsh, K.; O’Driscoll, M.; Longhurst, P.J.; Pollard, S.J.T.; Drew, G.H. Use of dispersion modelling for Environmental Impact Assessment of biological air pollution from composting: Progress, problems and prospects. Waste Manag. 2017. [Google Scholar] [CrossRef] [PubMed]
- Wéry, N. Bioaerosols from composting facilities—A review. Front. Cell. Infect. Microbiol. 2014, 4, 1–9. [Google Scholar]
- Douglas, P.; Bakolis, I.; Fecht, D.; Pearson, C.; Leal Sanchez, M.; Kinnersley, R.; de Hoogh, K.; Hansell, A.L. Respiratory hospital admission risk near large composting facilities. Int. J. Hyg. Environ. Health 2016, 219, 372–379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, K.; Kabir, E.; Jahan, S.A. Airborne bioaerosols and their impact on human health. J. Environ. Sci. 2017, 1–13. [Google Scholar] [CrossRef]
- Cerda, A.; Artola, A.; Font, X.; Barrena, R.; Gea, T.; Sánchez, A. Composting of food wastes: Status and challenges. Bioresour. Technol. 2018, 248, 57–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Douwes, J.; Thorne, P.; Pearce, N.; Heederik, D. Bioaerosol health effects and exposure assessment: Progress and prospects. Ann. Occup. Hyg. 2003, 47, 187–200. [Google Scholar]
- Douwes, J.; Travier, N.; Huang, K.; Cheng, S.; McKenzie, J.; Le Gros, G.; Von Mutius, E.; Pearce, N. Lifelong farm exposure may strongly reduce the risk of asthma in adults. Allergy Eur. J. Allergy Clin. Immunol. 2007, 62, 1158–1165. [Google Scholar] [CrossRef] [PubMed]
- Hoffmeyer, F.; van Kampen, V.; Taeger, D.; Deckert, A.; Rosenkranz, N.; Kaßen, M.; Schantora, A.; Brüning, T.; Raulf, M.; Bünger, J. Prevalence of and relationship between rhinoconjunctivitis and lower airway diseases in compost workers with current or former exposure to organic dust. Ann. Agric. Environ. Med. 2014, 21, 705–711. [Google Scholar] [CrossRef] [Green Version]
- Ho Man, W.; de Steenhuijsen Piters, W.A.A.; Bogaert, D. The microbiota of the respiratory tract: gatekeeper to respiratory health. Nat. Rev. Microbiol. 2017, 15, 259–270. [Google Scholar] [CrossRef] [PubMed]
- Eduard, W.; Heederik, D.; Duchaine, C.; Green, B.J. Bioaerosol exposure assessment in the workplace: The past, present and recent advances. J. Environ. Monit. 2012, 14, 334. [Google Scholar] [CrossRef]
- Pearson, C.; Littlewood, E.; Douglas, P.; Robertson, S.; Gant, T.W.; Hansell, A.L. Exposures and Health Outcomes in Relation to Bioaerosol Emissions From Composting Facilities: A Systematic Review of Occupational and Community Studies. J. Toxicol. Environ. Heal. Part B 2015, 18, 43–69. [Google Scholar] [CrossRef] [PubMed]
- Douglas, P.; Robertson, S.; Gay, R.; Hansell, A.L.; Gant, T.W. A systematic review of the public health risks of bioaerosols from intensive farming. Int. J. Hyg. Environ. Health 2018, 221, 134–173. [Google Scholar] [CrossRef] [PubMed]
- Walser, S.M.; Gerstner, D.G.; Brenner, B.; Bünger, J.; Eikmann, T.; Janssen, B.; Kolb, S.; Kolk, A.; Nowak, D.; Raulf, M.; et al. Evaluation of exposure-response relationships for health effects of microbial bioaerosols—A systematic review. Int. J. Hyg. Environ. Health 2015, 218, 577–589. [Google Scholar] [CrossRef] [PubMed]
- Mbareche, H.; Morawska, L.; Duchaine, C. Opinion paper on the challenges and proposed avenues for a better interpretation of bioaerosol exposure measurements and impacts on health. J. Air Waste Manag. Assoc. 2019, 7, 789–804. [Google Scholar]
- Le Goff, O.; Godon, J.J.; Milferstedt, K.; Bacheley, H.; Steyer, J.P.; Wéry, N. A new combination of microbial indicators for monitoring composting bioaerosols. Atmos. Environ. 2012, 61, 428–433. [Google Scholar] [CrossRef]
- Degois, J.; Clerc, F.; Simon, X.; Bontemps, C.; Leblond, P.; Duquenne, P. First Metagenomic Survey of the Microbial Diversity in Bioaerosols Emitted in Waste Sorting Plants. Ann. Work Expo. Heal. 2017, 61, 1076–1086. [Google Scholar] [CrossRef] [PubMed]
- Mubareka, S.; Groulx, N.; Savory, E.; Cutts, T.; Theriault, S.; Scott, J.A.; Roy, C.J.; Turgeon, N.; Bryce, E.; Astrakianakis, G.; et al. Bioaerosols and Transmission, a Diverse and Growing Community of Practice. Front. Public Heal. 2019, 7. [Google Scholar] [CrossRef]
- Traversi, D.; Gorrasi, I.; Pignata, C.; Degan, R.; Anedda, E.; Carletto, G.; Vercellino, G.; Fornasero, S.; Bertino, A.; Filippi, F.; et al. Aerosol exposure and risk assessment for green jobs involved in biomethanization. Environ. Int. 2018, 114, 202–211. [Google Scholar] [CrossRef]
- UNI. UNI CEN/TS 16115-1 Misurazione di Bioaerosol. Parte 1: Determinazione di Muffe, Utilizzando Sistemi di Campionamento di Filtrazione e Coltivazione; Centro Formazione UNI: Milano, Italy, 2011. [Google Scholar]
- ACGIH. 2018 Guide to Occupational Exposure Values; Governmental American Conference of Industrial Hygienists, Ed.; ACGIH: Cincinnati, OH, USA, 2018; ISBN 978-1-607260-98-1. [Google Scholar]
- Zhao, Y.; Aarnink, A.; Wang, W.; Fabri, T.; Groot Koerkamp, P.W.G.; de Jong, M.C.M. Airborne virus sampling: Efficiencies of samplers and their detection limits for infectious bursal disease virus (IBDV). Ann. Agric. Environ. Med. 2014, 21, 464–471. [Google Scholar] [CrossRef] [Green Version]
- Ziros, P.G.; Kokkinos, P.A.; Legaki, E.; Vantarakis, A. Development of an optimized method for the detection of airborne viruses with real-time PCR analysis. Virol. J. 2011, 8, 369. [Google Scholar] [CrossRef]
- Farell, E.M.; Alexandre, G. Bovine serum albumin further enhances the effects of organic solvents on increased yield of polymerase chain reaction of GC-rich templates. BMC Res. Notes 2012, 5, 257. [Google Scholar] [CrossRef]
- Muyzer, G.; de Waal, E.C.; Uitterlinden, A.G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 1993, 59, 695–700. [Google Scholar] [PubMed]
- O’Sullivan, L.A.; Webster, G.; Fry, J.C.; Parkes, R.J.; Weightman, A.J. Modified linker-PCR primers facilitate complete sequencing of DGGE DNA fragments. J. Microbiol. Methods 2008, 75, 579–581. [Google Scholar] [CrossRef] [PubMed]
- Dridi, B.; Henry, M.; El Khéchine, A.; Raoult, D.; Drancourt, M. High prevalence of Methanobrevibacter smithii and Methanosphaera stadtmanae detected in the human gut using an improved DNA detection protocol. PLoS ONE 2009, 4. [Google Scholar] [CrossRef]
- Yoo, K.; Lee, T.K.; Choi, E.J.; Yang, J.; Shukla, S.K.; Hwang, S.I.; Park, J. Molecular approaches for the detection and monitoring of microbial communities in bioaerosols: A review. J. Environ. Sci. 2017, 51, 234–247. [Google Scholar] [CrossRef]
- Kim, Y.M.; Jang, H.M.; Lee, K.; Chantrasakdakul, P.; Kim, D.; Park, K.Y. Changes in bacterial and archaeal communities in anaerobic digesters treating different organic wastes. Chemosphere 2015, 141, 134–137. [Google Scholar] [CrossRef] [PubMed]
- Trzcinski, A.P.; Ray, M.J.; Stuckey, D.C. Performance of a three-stage membrane bioprocess treating the organic fraction of municipal solid waste and evolution of its archaeal and bacterial ecology. Bioresour. Technol. 2010, 101, 1652–1661. [Google Scholar] [CrossRef]
- Chakravorty, S.; Helb, D.; Burday, M.; Connell, N.; Alland, D. A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. J. Microbiol. Methods 2007, 69, 330–339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flemming, H.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010. [Google Scholar] [CrossRef]
- Simões, M.; Simões, L.C.; Vieira, M.J. Species association increases biofilm resistance to chemical and mechanical treatments. Water Res. 2009, 43, 229–237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- European Centre for Disease Prevention and Control. Surveillance of Antimicrobial Resistance in Europe 2016. Annual Report of the European Antimicrobial Resistance Surveillance Network; European Centre for Disease Prevention and Control: Solna, Sweden, 2017; ISBN 9789294980991. [Google Scholar]
- Stiles, B.G.; Pradhan, K.; Fleming, J.M.; Samy, R.P.; Barth, H.; Popoff, M.R. Clostridium and bacillus binary enterotoxins: bad for the bowels, and eukaryotic being. Toxins 2014, 6, 2626–2656. [Google Scholar] [CrossRef]
- Sumampouw, O.J.; Risjani, Y. Bacteria as indicators of environmental pollution: Review. Int. J. Ecosyst. 2014, 4, 251–258. [Google Scholar]
- Ferreira, S.; Queiroz, J.A.; Oleastro, M.; Domingues, F.C. Insights in the pathogenesis and resistance of Arcobacter: A review. Crit. Rev. Microbiol. 2016, 42, 364–383. [Google Scholar] [PubMed]
- Resende, J.A.; Silva, V.L.; de Oliveira, T.L.R.; de Oliveira Fortunato, S.; da Costa Carneiro, J.; Otenio, M.H.; Diniz, C.G. Prevalence and persistence of potentially pathogenic and antibiotic resistant bacteria during anaerobic digestion treatment of cattle manure. Bioresour. Technol. 2014, 153, 284–291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aerts, R.; Honnay, O.; Nieuwenhuyse, A. Van Biodiversity and human health: mechanisms and evidence of the positive health effects of diversity in nature and green spaces. Br. Med. Bull. 2018, 127. [Google Scholar] [CrossRef]



| Plant Type | N° Filters | N° LMA Plates |
|---|---|---|
| OFMSW | 5 | 4 |
| ALB | 17 | 12 |
| WWTS | 4 | 4 |
| LCB | 6 | 4 |
| Primers | Sequence (5′−3′) | Target Genes | References |
|---|---|---|---|
| 357F-GCa | GCclamp -CTC CTA CGG GAG GCA GCA G | 16S rRNA Bacteria | [25] |
| 518R | GTA TTA CCG CGG CTG CTG G | ||
| 357F-GC | GCclamp -CTC CTA CGG GAG GCA GCA G | 16S rRNA Bacteria | [25,26] |
| 518R-AT-M13 | GTAAAACGACGGCCAGTAAATAAAATAAAAATGTAAAAAAATTACCGCGGCTGCTGG | ||
| TotBact F TotBact R | ACTCCTACGGGAGGCAGCAG ATTACCGCGGCTGCTGG | 16S rDNA Total Bacteria | [27] |
| Plant Type | LMA Bacterial Count Log10 (gene copies/m3) | Total Count at 37 °C Log10 (UFC/m3) | Filter Bacterial Count Log10 (gene copies/m3) |
|---|---|---|---|
| ALB | 4.27 ± 0.35 | 2.49 ± 0.44 | 1.65 ± 0.16 |
| LCB | 4.75 ± 0.20 | 3.14 ± 0.97 | 3.03 ± 0.33 |
| OFMSW | 4.41 ± 0.31 | 3.18 ± 0.97 | 2.42 ± 0.86 |
| WWTP | 3.76 ± 0.35 | 2.36 ± 1.23 | 1.60 ± 0.07 |
| Plants | Area | Length | Closest Relative | Accession N° | % | Phylum | Order | Family | Genus |
|---|---|---|---|---|---|---|---|---|---|
| OFMWS | Input | 160 | Enterobacter sp. | GQ465229 | 95 | Proteobacteria | Enterobacterales | Enterobacteriaceae | Enterobacter |
| 159 | Enterobacter cloacae | KX303810 | 98 | Proteobacteria | Enterobacterales | Enterobacteriaceae | Enterobacter | ||
| 160 | Uncultured Bacillus sp. | KM819125 | 94 | Firmicutes | Bacillales | Bacillaceae | Bacillus | ||
| 158 | Uncultured Enterococcus sp. | LT625552 | 91 | Firmicutes | Lactobacillales | Enterococcaceae | Enterococcus | ||
| 160 | Lactobacillus sp. | KC755084 | 100 | Firmicutes | Lactobacillales | Lactobacillaceae | Lactobacillus | ||
| 158 | Uncultured Klebsiella sp. | LC342860 | 90 | Proteobacteria | Enterobacterales | Enterobacteriaceae | Klebsiella | ||
| Output | 135 | Arcobacter skirrowii | MG195899 | 100 | Proteobacteria | Campylobacterales | Campylobacteraceae | Arcobacter | |
| 160 | Pseudomonas fluorescens | EU434416 | 94 | Proteobacteria | Pseudomonales | Pseudomonadaceae | Pseudomonas | ||
| 160 | Pseudomonas protegens | MG269715 | 100 | Proteobacteria | Pseudomonales | Pseudomonadaceae | Pseudomonas | ||
| 160 | Comamonas sp. | JF928565 | 100 | Proteobacteria | Burkholderiales | Comamonadaceae | Comamonas | ||
| ALB | Output | 160 | Uncultured Ralstonia sp. | MG801673 | 100 | Proteobacteria | Burkholderiales | Burkholderiaceae | Ralstonia |
| WWTS | Input | 160 | Comamonas sp. | JF928565 | 100 | Proteobacteria | Burkholderiales | Comamonadaceae | Comamonas |
| 160 | Uncultured Ralstonia sp. | MG801673 | 98 | Proteobacteria | Burkholderiales | Burkholderiaceae | Ralstonia | ||
| LBC | Input | 160 | Pseudomonas sp. | MH394449 | 100 | Proteobacteria | Pseudomonales | Pseudomonadaceae | Pseudomonas |
| Output | 135 | Arcobacter skirrowii | MG195900 | 100 | Proteobacteria | Campylobacterales | Campylobacteraceae | Arcobacter |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anedda, E.; Carletto, G.; Gilli, G.; Traversi, D. Monitoring of Air Microbial Contaminations in Different Bioenergy Facilities Using Cultural and Biomolecular Methods. Int. J. Environ. Res. Public Health 2019, 16, 2546. https://doi.org/10.3390/ijerph16142546
Anedda E, Carletto G, Gilli G, Traversi D. Monitoring of Air Microbial Contaminations in Different Bioenergy Facilities Using Cultural and Biomolecular Methods. International Journal of Environmental Research and Public Health. 2019; 16(14):2546. https://doi.org/10.3390/ijerph16142546
Chicago/Turabian StyleAnedda, Elisa, Giulia Carletto, Giorgio Gilli, and Deborah Traversi. 2019. "Monitoring of Air Microbial Contaminations in Different Bioenergy Facilities Using Cultural and Biomolecular Methods" International Journal of Environmental Research and Public Health 16, no. 14: 2546. https://doi.org/10.3390/ijerph16142546
APA StyleAnedda, E., Carletto, G., Gilli, G., & Traversi, D. (2019). Monitoring of Air Microbial Contaminations in Different Bioenergy Facilities Using Cultural and Biomolecular Methods. International Journal of Environmental Research and Public Health, 16(14), 2546. https://doi.org/10.3390/ijerph16142546