Ruminant Fecal Contamination of Drinking Water Introduced Post-Collection in Rural Kenyan Households
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Site and Sample Frame
2.2. Fecal Sample Collection and Processing
2.3. Household Water and Survey Data Collection
2.4. Molecular Processing of Fecal and Environmental Samples
2.5. MST Validation Data Analysis
2.6. Data Analysis—Water Samples
2.7. fsQCA
3. Results
3.1. Quality Assurance/Quality Control
3.2. MST Validation Study
3.3. Household Characteristics
3.4. Animal Characteristics and Host-Associated MST Marker Results
3.5. Water Supply Characteristics
3.6. Ruminant Contamination Introduced to Stored Water
3.7. Correlations Between Fecal Indicators
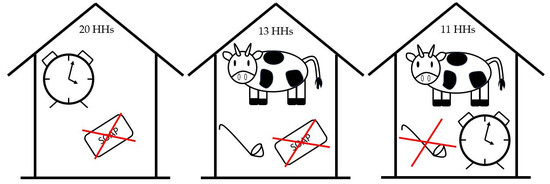
3.8. Combinations for Introduction of Ruminant Contamination to Stored Water Samples
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
Appendix B
| Ruminants | Unsafe Extraction | Long Storage Time | No Soap | Number | Raw Consist. | PRI Consist. | SYM Consist. |
|---|---|---|---|---|---|---|---|
| 1 | 1 | 1 | 1 | 11 (33%) | 0.943434 | 0.943434 | 0.943434 |
| 1 | 0 | 1 | 1 | 9 (60%) | 0.885057 | 0.885057 | 0.885057 |
| 0 | 1 | 1 | 1 | 3 (69%) | 0.935058 | 0.935058 | 0.935058 |
| 1 | 0 | 1 | 0 | 2 (75%) | 1 | 1 | 1 |
| 1 | 1 | 1 | 0 | 2 (81%) | 0.5 | 0.5 | 0.5 |
| 0 | 1 | 0 | 1 | 2 (87%) | 0.438849 | 0.438849 | 0.438849 |
| 1 | 1 | 0 | 1 | 2 (93%) | 0.858064 | 0.858064 | 0.858065 |
| 0 | 0 | 1 | 0 | 1 (97%) | 0 | 0 | 0 |
| 0 | 0 | 1 | 1 | 1 (100%) | 1 | 1 | 1 |
| 0 | 0 | 0 | 0 | 0 (100%) | |||
| 1 | 0 | 0 | 0 | 0 (100%) | |||
| 0 | 1 | 0 | 0 | 0 (100%) | |||
| 1 | 1 | 0 | 0 | 0 (100%) | |||
| 0 | 1 | 1 | 0 | 0 (100%) | |||
| 0 | 0 | 0 | 1 | 0 (100%) | |||
| 1 | 0 | 0 | 1 | 0 (100%) |

Appendix C
| Avians Present | Avians Absent | |
|---|---|---|
| Avian GFD Detected | 2 | 0 |
| Avian GFD Not Detected | 37 | 6 |
| Ruminants Present | Ruminants Absent | |
|---|---|---|
| BacR Detected | 31 | 9 |
| BacR Not Detected | 3 | 2 |
Appendix D

References
- WHO/UNICEF Joint Water Supply, and Sanitation Monitoring Programme. Progress on Drinking Water and Sanitation: 2014 Update; WHO: Geneva, Switzerland, 2014; Available online: https://www.who.int/water_sanitation_health/publications/2014/jmp-report/en/ (accessed on 15 January 2020).
- JMP. Progress on Drinking Water, Sanitation and Hygiene; JMP: Geneva, Switzerland, 2017; Available online: http://www.who.int/water_sanitation_health/publications/jmp-2017/en/ (accessed on 15 January 2020).
- Wright, J.; Gundry, S.; Conroy, R. Household drinking water in developing countries: A systematic review of microbiological contamination between source and point-of-use. Trop. Med. Int. Health 2004, 9, 106–117. [Google Scholar] [CrossRef] [Green Version]
- Brick, T.; Primrose, B.; Chandrasekhar, R.; Roy, S.; Muliyil, J.; Kang, G. Water contamination in urban south India: Household storage practices and their implications for water safety and enteric infections. Int. J. Hyg. Environ. Health 2004, 207, 473–480. [Google Scholar] [CrossRef]
- Mølbak, K.; Højlyng, N.; Jepsen, S.; Gaarslev, K. Bacterial contamination of stored water and stored food: A potential source of diarrhoeal disease in West Africa. Epidemiol. Infect. 1989, 102, 309–316. [Google Scholar] [CrossRef] [Green Version]
- Schriewer, A.; Odagiri, M.; Wuertz, S.; Misra, P.R.; Panigrahi, P.; Clasen, T.; Jenkins, M.W. Human and Animal Fecal Contamination of Community Water Sources, Stored Drinking Water and Hands in Rural India Measured with Validated Microbial Source Tracking Assays. Am. J. Trop. Med. Hyg. 2015, 93, 509–516. [Google Scholar] [CrossRef]
- Pickering, A.J.; Davis, J.; Walters, S.P.; Horak, H.M.; Keymer, D.P.; Mushi, D.; Strickfaden, R.; Chynoweth, J.S.; Liu, J.; Blum, A.; et al. Hands, Water, and Health: Fecal Contamination in Tanzanian Communities with Improved, Non-Networked Water Supplies. Environ. Sci. Technol. 2010, 44, 3267–3272. [Google Scholar] [CrossRef]
- Ercumen, A.; Naser, A.M.; Unicomb, L.; Arnold, B.F.; Colford, J.; Luby, S.P. Effects of Source—Versus Household Contamination of Tubewell Water on Child Diarrhea in Rural Bangladesh: A Randomized Controlled Trial. PLoS ONE 2015, 10, e0121907. [Google Scholar] [CrossRef]
- Hunter, P.R.; Zmirou-Navier, D.; Hartemann, P. Estimating the impact on health of poor reliability of drinking water interventions in developing countries. Sci Total Environ. 2009, 407, 2621–2624. [Google Scholar] [CrossRef]
- Harris, A.R.; Davis, J.; Boehm, A.B. Mechanisms of post-supply contamination of drinking water in Bagamoyo, Tanzania. J. Water Health 2013, 11, 543–554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oswald, W.E.; Lescano, A.G.; Bern, C.; Calderon, M.M.; Cabrera, L.; Gilman, R.H. Fecal Contamination of Drinking Water within Peri-Urban Households, Lima, Peru. Am. J. Trop. Med. Hyg. 2007, 77, 699–704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boehm, A.B.; Wang, D.; Ercumen, A.; Shea, M.; Harris, A.R.; Shanks, O.C.; Kelty, C.; Ahmed, A.; Mahmud, Z.H.; Arnold, B.F.; et al. Occurrence of Host-Associated Fecal Markers on Child Hands, Household Soil, and Drinking Water in Rural Bangladeshi Households. Environ. Sci. Technol. Lett. 2016, 3, 393–398. [Google Scholar] [CrossRef]
- Harris, A.R.; Pickering, A.J.; Boehm, A.B.; Mrisho, M.; Davis, J. Comparison of analytical techniques to explain variability in stored drinking water quality and microbial hand contamination of female caregivers in Tanzania. Environ. Sci. Process. Impacts 2019, 21, 893–903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andreas, J.J.; Burns, C.; Touza, J. Renewable Energy as a Luxury? A Qualitative Comparative Analysis of the Role of the Economy in the EU’s Renewable Energy Transitions During the ‘Double Crisis.’ Ecol. Econom. 2017, 142, 81–90. [Google Scholar]
- Pappas, I.O.; Kourouthanassis, P.E.; Giannakos, M.N.; Chrissikopoulos, V. Explaining online shopping behavior with fsQCA: The role of cognitive and affective perceptions. J. Bus. Res. 2016, 69, 794–803. [Google Scholar] [CrossRef]
- Oyemomi, O.; Liu, S.; Neaga, I.; Alkhuraiji, A. How knowledge sharing and business process contribute to organizational performance: Using the fsQCA approach. J. Bus. Res. 2016, 69, 5222–5227. [Google Scholar] [CrossRef] [Green Version]
- Marks, S.J.; Kumpel, E.; Guo, J.; Bartram, J.; Davis, J. Pathways to sustainability: A fuzzy-set qualitative comparative analysis of rural water supply programs. J. Clean. Prod. 2018, 205, 789–798. [Google Scholar] [CrossRef]
- World Health Organisation. Guidelines for Drinking-Water Quality, 3rd ed.; World Health Organisation: Geneva, Switzerland, 2008; Volume 1, Available online: http://www.who.int/water_sanitation_health/publications/gdwq3rev/en/ (accessed on 15 January 2020).
- Bartram, J.; Rees, G. Monitoring Bathing Waters: A Practical Guide to the Design and Implementation of Assessments and Monitoring Programmes; CRC Press: Boca Raton, FL, USA, 2002; Available online: https://www.taylorfrancis.com/books/9781135920661 (accessed on 15 January 2020).
- Ashbolt, N.J. Microbial contamination of drinking water and disease outcomes in developing regions. Toxicology 2004, 198, 229–238. [Google Scholar] [CrossRef] [PubMed]
- Edberg, S.C.; Rice, E.W.; Karlin, R.J.; Allen, M.J. Escherichia coli: The best biological drinking water indicator for public health protection. J. Appl. Microbiol. 2000, 88, 106S–116S. [Google Scholar] [CrossRef]
- Lemarchand, K.; Lebaron, P. Occurrence of Salmonella spp and Cryptosporidium spp in a French coastal watershed: Relationship with fecal indicators. FEMS Microbiol. Lett. 2003, 218, 203–209. [Google Scholar] [CrossRef] [Green Version]
- Lund, V. Evaluation of E. coli as an indicator for the presence of Campylobacter jejuni and Yersinia enterocolitica in chlorinated and untreated oligotrophic lake water. Water Res. 1996, 30, 1528–1534. [Google Scholar] [CrossRef]
- Knappett, P.S.; McKay, L.D.; Layton, A.; Williams, D.E.; Alam, M.J.; Mailloux, B.J.; Ferguson, A.S.; Culligan, P.J.; Serre, M.L.; Emch, M.; et al. Unsealed tubewells lead to increased fecal contamination of drinking water. J. Water Health 2012, 10, 565–578. [Google Scholar] [CrossRef]
- Tallon, P.; Magajna, B.; Lofranco, C.; Leung, K.T. Microbial Indicators of Faecal Contamination in Water: A Current Perspective. Water Air Soil Pollut. 2005, 166, 139–166. [Google Scholar] [CrossRef]
- Verhille, S. Understanding Microbial Indicators for Drinking Water Assessment: Interpretation of Test Results and Public Health Significance. National Collaborating Centre for Environmental Health. 2013. Available online: http://www.ncceh.ca/sites/default/files/Microbial_Indicators_Jan_2013_0.pdf (accessed on 15 January 2020).
- Soller, J.A.; Schoen, M.E.; Bartrand, T.; Ravenscroft, J.E.; Ashbolt, N.J. Estimated human health risks from exposure to recreational waters impacted by human and non-human sources of faecal contamination. Water Res. 2010, 44, 4674–4691. [Google Scholar] [CrossRef] [PubMed]
- Bernhard, A.E.; Field, K.G. A PCR Assay to Discriminate Human and Ruminant Feces on the Basis of Host Differences in Bacteroides-Prevotella Genes Encoding 16S rRNA. Appl. Environ. Microbiol. 2000, 66, 4571–4574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scott, T.M.; Rose, J.B.; Jenkins, T.M.; Farrah, S.R.; Lukasik, J. Microbial Source Tracking: Current Methodology and Future Directions. Appl. Environ. Microbiol. 2002, 68, 5796–5803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parker, J.K.; McIntyre, D.; Noble, R.T. Characterizing fecal contamination in stormwater runoff in coastal North Carolina, USA. Water Res. 2010, 44, 4186–4194. [Google Scholar] [CrossRef] [PubMed]
- Santo Domingo, J.W.; Bambic, D.G.; Edge, T.A.; Wuertz, S. Quo vadis source tracking? Towards a strategic framework for environmental monitoring of fecal pollution. Water Res. 2007, 41, 3539–3552. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Yusuf, R.; Hasan, I.; Goonetilleke, A.; Gardner, T. Quantitative PCR assay of sewage-associated Bacteroides markers to assess sewage pollution in an urban lake in Dhaka, Bangladesh. Can. J. Microbiol. 2010, 56, 838–845. [Google Scholar] [CrossRef] [Green Version]
- Jenkins, M.W.; Tiwari, S.; Lorente, M.; Gichaba, C.M.; Wuertz, S. Identifying human and livestock sources of fecal contamination in Kenya with host-specific Bacteroidales assays. Water Res. 2009, 43, 4956–4966. [Google Scholar] [CrossRef]
- Odagiri, M.; Schriewer, A.; Hanley, K.; Wuertz, S.; Misra, P.R.; Panigrahi, P.; Jenkins, M.W. Validation of Bacteroidales quantitative PCR assays targeting human and animal fecal contamination in the public and domestic domains in India. Sci. Total. Environ. 2015, 502, 462–470. [Google Scholar] [CrossRef] [Green Version]
- Reischer, G.H.; Ebdon, J.E.; Bauer, J.M.; Schuster, N.; Ahmed, W.; Åström, J.; Blanch, A.R.; Blöschl, G.; Byamukama, D.; Coakley, T.; et al. Performance Characteristics of qPCR Assays Targeting Human- and Ruminant-Associated Bacteroidetes for Microbial Source Tracking across Sixteen Countries on Six Continents. Environ. Sci. Technol. 2013, 47, 8548–8556. [Google Scholar] [CrossRef]
- Salah, D.M.M.A.; Laffite, A.; Poté, J. Occurrence of Bacterial Markers and Antibiotic Resistance Genes in Sub-Saharan Rivers Receiving Animal Farm Wastewaters. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Odagiri, M.; Schriewer, A.; Daniels, M.E.; Wuertz, S.; Smith, W.A.; Clasen, T.; Schmidt, W.P.; Jin, Y.; Torondel, B.; Misra, P.R.; et al. Human fecal and pathogen exposure pathways in rural Indian villages and the effect of increased latrine coverage. Water Res. 2016, 100, 232–244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pickering, A.J.; Njenga, S.M.; Steinbaum, L.; Swarthout, J.; Lin, A.; Arnold, B.F.; Stewart, C.P.; Dentz, H.N.; Mureithi, M.; Chieng, B.; et al. Effects of single and integrated water, sanitation, handwashing, and nutrition interventions on child soil-transmitted helminth and Giardia infections: A cluster-randomized controlled trial in rural Kenya. PLOS Med. 2019, 16, e1002841. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Null, C.; Stewart, C.P.; Pickering, A.J.; Dentz, H.N.; Arnold, B.F.; Arnold, C.D.; Benjamin-Chung, J.; Clasen, T.; Dewey, K.D.; Fernald, L.C.H.; et al. Effects of water quality, sanitation, handwashing, and nutritional interventions on diarrhoea and child growth in rural Kenya: A cluster-randomised controlled trial. Lancet Glob. Health 2018, 6, e316–e329. [Google Scholar] [CrossRef] [Green Version]
- Pickering, A.J.; Null, C.; Winch, P.J.; Mangwadu, G.; Arnold, B.F.; Prendergast, A.J.; Njenga, S.M.; Rahman, M.; Ntozini, R.; Benjamin-Chung, J.; et al. The WASH Benefits and SHINE trials: Interpretation of WASH intervention effects on linear growth and diarrhoea. Lancet Glob. Health 2019, 7, e1139–e1146. [Google Scholar] [CrossRef] [Green Version]
- Arnold, B.F.; Null, C.; Luby, S.P.; Unicomb, L.; Stewart, C.P.; Dewey, K.G.; Ahmed, T.; Ashraf, S.; Christensen, G.; Clasen, T.; et al. Cluster-randomised controlled trials of individual and combined water, sanitation, hygiene and nutritional interventions in rural Bangladesh and Kenya: The WASH Benefits study design and rationale. BMJ Open 2013, 3, e003476. [Google Scholar] [CrossRef] [Green Version]
- Pickering, A.J.; Arnold, B.F.; Dentz, H.N.; Colford, J.M.; Null, C. Climate and Health Co-Benefits in Low-Income Countries: A Case Study of Carbon Financed Water Filters in Kenya and a Call for Independent Monitoring. Environ. Health Perspect. 2017, 125, 278–283. [Google Scholar] [CrossRef] [Green Version]
- Harris, A.R. Understanding Sources and Determinants of Fecal Contamination of Water, Hands, Food, and Household Floors in Low-Income Countries. Ph.D. Thesis, Stanford University, Stanford, CA, USA, June 2015. [Google Scholar]
- US Environmental Protection Agency. Method 1604: Total Coliforms and Escherichia coli in Water by Membrane Filtration Using a Simultaneous Detection Technique (MI Medium); US Environmental Protection Agency, 2002. Available online: https://nepis.epa.gov/Exe/ZyNET.exe/P1002D57.TXT?ZyActionD=ZyDocument&Client=EPA&Index=2000+Thru+2005&Docs=&Query=&Time=&EndTime=&SearchMethod=1&TocRestrict=n&Toc=&TocEntry=&QField=&QFieldYear=&QFieldMonth=&QFieldDay=&IntQFieldOp=0&ExtQFieldOp=0&XmlQuery=&File=D%3A%5Czyfiles%5CIndex%20Data%5C00thru05%5CTxt%5C00000017%5CP1002D57.txt&User=ANONYMOUS&Password=anonymous&SortMethod=h%7C-&MaximumDocuments=1&FuzzyDegree=0&ImageQuality=r75g8/r75g8/x150y150g16/i425&Display=hpfr&DefSeekPage=x&SearchBack=ZyActionL&Back=ZyActionS&BackDesc=Results%20page&MaximumPages=1&ZyEntry=1&SeekPage=x&ZyPURL (accessed on 15 January 2020).
- US Environmental Protection Agency. Method 1600: Enterococci in Water by Membrane Filtration Using membrane- Enterococcus Indoxyl-$-D-Glucoside Agar (mEI). 2006. Available online: https://nepis.epa.gov/Exe/ZyNET.exe/P10099DH.TXT?ZyActionD=ZyDocument&Client=EPA&Index=2000+Thru+2005&Docs=&Query=&Time=&EndTime=&SearchMethod=1&TocRestrict=n&Toc=&TocEntry=&QField=&QFieldYear=&QFieldMonth=&QFieldDay=&IntQFieldOp=0&ExtQFieldOp=0&XmlQuery=&File=D%3A%5Czyfiles%5CIndex%20Data%5C00thru05%5CTxt%5C00000025%5CP10099DH.txt&User=ANONYMOUS&Password=anonymous&SortMethod=h%7C-&MaximumDocuments=1&FuzzyDegree=0&ImageQuality=r75g8/r75g8/x150y150g16/i425&Display=hpfr&DefSeekPage=x&SearchBack=ZyActionL&Back=ZyActionS&BackDesc=Results%20page&MaximumPages=1&ZyEntry=1&SeekPage=x&ZyPURL (accessed on 15 January 2020).
- Harris, A.R.; Pickering, A.J.; Harris, M.; Doza, S.; Islam, M.S.; Unicomb, L.; Luby, S.; Davis, J.; Boehm, A.B. Ruminants Contribute Fecal Contamination to the Urban Household Environment in Dhaka, Bangladesh. Environ. Sci. Technol. 2016, 50, 4642–4649. [Google Scholar] [CrossRef]
- Haugland, R.A.; Varma, M.; Sivaganesan, M.; Kelty, C.; Peed, L.; Shanks, O.C. Evaluation of genetic markers from the 16S rRNA gene V2 region for use in quantitative detection of selected Bacteroidales species and human fecal waste by qPCR. Syst. Appl. Microbiol. 2010, 33, 348–357. [Google Scholar] [CrossRef]
- Kildare, B.J.; Leutenegger, C.M.; McSwain, B.S.; Bambic, D.G.; Rajal, V.B.; Wuertz, S. 16S rRNA-based assays for quantitative detection of universal, human-, cow-, and dog-specific fecal Bacteroidales: A Bayesian approach. Water Res. 2007, 41, 3701–3715. [Google Scholar] [CrossRef]
- Shanks, O.C.; Kelty, C.A.; Sivaganesan, M.; Varma, M.; Haugland, R.A. Quantitative PCR for Genetic Markers of Human Fecal Pollution. Appl. Environ. Microbiol. 2009, 75, 5507–5513. [Google Scholar] [CrossRef] [Green Version]
- Mieszkin, S.; Yala, J.F.; Joubrel, R.; Gourmelon, M. Phylogenetic analysis of Bacteroidales 16S rRNA gene sequences from human and animal effluents and assessment of ruminant faecal pollution by real-time PCR. J. Appl. Microbiol. 2010, 108, 974–984. [Google Scholar] [CrossRef] [Green Version]
- Reischer, G.H.; Kasper, D.C.; Steinborn, R.; Mach, R.L.; Farnleitner, A.H. Quantitative PCR Method for Sensitive Detection of Ruminant Fecal Pollution in Freshwater and Evaluation of This Method in Alpine Karstic Regions. Appl. Environ. Microbiol. 2006, 72, 5610–5614. [Google Scholar] [CrossRef] [Green Version]
- Green, H.C.; Dick, L.K.; Gilpin, B.; Samadpour, M.; Field, K.G. Genetic Markers for Rapid PCR-Based Identification of Gull, Canada Goose, Duck, and Chicken Fecal Contamination in Water. Appl. Environ. Microbiol. 2012, 78, 503–510. [Google Scholar] [CrossRef] [Green Version]
- Boehm, A.B.; Van De Werfhorst, L.C.; Griffith, J.F.; Holden, P.A.; Jay, J.A.; Shanks, O.C.; Wang, D.; Weisberg, S.B. Performance of forty-one microbial source tracking methods: A twenty-seven lab evaluation study. Water Res. 2013, 47, 6812–6828. [Google Scholar] [CrossRef]
- Rihoux, B.; Ragin, C. Configurational Comparative Methods: Qualitative Comparative Analysis (QCA) and Related Techniques; SAGE Publications, Inc.: Thousand Oaks, CA, USA, 2009; Available online: http://methods.sagepub.com/book/configurational-comparative-methods (accessed on 15 January 2020).
- Mahoney, J.; Goertz, G. The Possibility Principle: Choosing Negative Cases in Comparative Research. Am. Political Sci. Rev. 2004, 98, 653–669. [Google Scholar] [CrossRef]
- Penakalapati, G.; Swarthout, J.; Delahoy, M.J.; McAliley, L.; Wodnik, B.; Levy, K.; Freeman, M.C. Exposure to Animal Feces and Human Health: A Systematic Review and Proposed Research Priorities. Environ. Sci Technol. 2017, 51, 11537–11552. [Google Scholar] [CrossRef] [Green Version]
- Fayer, R.; Dubey, J.P.; Lindsay, D.S. Zoonotic protozoa: From land to sea. Trends Parasitol. 2004, 20, 531–536. [Google Scholar] [CrossRef]
- Delahoy, M.J.; Wodnik, B.; McAliley, L.; Penakalapati, G.; Swarthout, J.; Freeman, M.C.; Lavy, K. Pathogens Transmitted in Animal Feces in Low- and Middle-Income Countries. Int. J. Hyg. Environ. Health 2018, 221, 661–676. Available online: http://www.sciencedirect.com/science/article/pii/S1438463917308635 (accessed on 15 January 2020). [CrossRef]
- Barnes, A.N.; Anderson, J.D.; Mumma, J.; Mahmud, Z.H.; Cumming, O. The association between domestic animal presence and ownership and household drinking water contamination among peri-urban communities of Kisumu, Kenya. PLoS ONE 2018, 13, e0197587. [Google Scholar] [CrossRef]
- Fuhrmeister, E.R.; Ercumen, A.; Pickering, A.J.; Jeanis, K.M.; Ahmed, M.; Brown, S.; Arnold, B.F.; Hubbard, A.E.; Alam, M.; Sen, D.; et al. Predictors of Enteric Pathogens in the Domestic Environment from Human and Animal Sources in Rural Bangladesh. Environ. Sci. Technol. 2019, 53, 10023–10033. [Google Scholar] [CrossRef] [Green Version]
- Curtis, V.; Cairncross, S. Effect of washing hands with soap on diarrhoea risk in the community: A systematic review. Lancet Infect. Dis. 2003, 3, 275–281. [Google Scholar] [CrossRef]
- Luby, S.P.; Agboatwalla, M.; Painter, J.; Altaf, A.; Billhimer, W.L.; Hoekstra, R.M. Effect of Intensive Handwashing Promotion on Childhood Diarrhea in High-Risk Communities in Pakistan: A Randomized Controlled Trial. JAMA 2004, 291, 2547–2554. [Google Scholar] [CrossRef]
- Dufour, A.; Bartram, J.; Bos, R.; Gannon, V. Animal Waste, Water Quality and Human Health. 2012. Available online: http://www.who.int/water_sanitation_health/publications/animal_waste/en/ (accessed on 15 January 2020).
- Prendergast, A.J.; Gharpure, R.; Mor, S.; Viney, M.; Dube, K.; Lello, J.; Berger, C.; Siwila, J.; Joyeux, M.; Hodobo, T.; et al. Putting the “A” into WaSH: A call for integrated management of water, animals, sanitation, and hygiene. Lancet Planet. Health 2019, 3, e336–e337. [Google Scholar] [CrossRef] [Green Version]
- Freeman, M.C.; Stocks, M.E.; Cumming, O.; Jeandron, A.; Higgins, J.P.T.; Wolf, J.; Prüss-Ustün, A.; Bonjour, S.; Hunter, P.R.; Fewtrell, L.; et al. Systematic review: Hygiene and health: Systematic review of handwashing practices worldwide and update of health effects. Trop. Med. Int. Health 2014, 19, 906–916. [Google Scholar] [CrossRef] [Green Version]
- Wolf, J.; Prüss-Ustün, A.; Cumming, O.; Bartram, J.; Bonjour, S.; Cairncross, S.; Clasen, T.; Colford, J.M.; Curtis, V.; De France, J.; et al. Systematic review: Assessing the impact of drinking water and sanitation on diarrhoeal disease in low- and middle-income settings: Systematic review and meta-regression. Trop. Med. Int. Health 2014, 19, 928–942. [Google Scholar] [CrossRef]
- Clasen, T.; Roberts, I.; Rabie, T.; Schmidt, W.; Cairncross, S. Interventions to improve water quality for preventing diarrhoea. Cochrane Database Syst. Rev. 2006, 3, CD004794. [Google Scholar]
- Fewtrell, L.; Kaufmann, R.B.; Kay, D.; Enanoria, W.; Haller, L.; Colford, J.M. Water, sanitation, and hygiene interventions to reduce diarrhoea in less developed countries: A systematic review and meta-analysis. Lancet Infect. Dis. 2005, 5, 42–52. [Google Scholar] [CrossRef]
- Waddington, H.; Snilstveit, B.; White, H.D.; Fewtrell, L. Water sanitation and hygiene interventions to combat childhood diarrhoea in developing countries. Synth. Rev. 2009, 3, 295–335. [Google Scholar]
- Kang’ethe, E.K.; Kimani, V.N.; McDermott, B.; Grace, D.; Lang’at, A.K.; Kiragu, M.W.; Karanja, N.; Njehu, A.N.; Randolph, T.; Mbugua, G.; et al. A trans-disciplinary study on the health risks of cryptosporidiosis from dairy systems in Dagoretti, Nairobi, Kenya: Study background and farming system characteristics. Trop. Anim. Health Prod. 2012, 44, 3–10. [Google Scholar] [CrossRef]
- Bokkers, E.A.M. Effects of Interactions between Humans and Domesticated Animals; Hassink, J., Van Dijk, M., Eds.; Springer: Dordrecht, The Netherlands, 2006; pp. 31–41. Available online: http://link.springer.com/10.1007/1-4020-4541-7_3 (accessed on 15 January 2020).
- Soest, P.J.V. Nutritional Ecology of the Ruminant; Cornell University Press: New York, NY, USA, 2018. [Google Scholar]
- Cao, Y.; Griffith, J.F.; Dorevitch, S.; Weisberg, S.B. Effectiveness of qPCR permutations, internal controls and dilution as means for minimizing the impact of inhibition while measuring Enterococcus in environmental waters. J. Appl. Microbiol. 2012, 113, 66–75. [Google Scholar] [CrossRef]




| Construct | Definition | Theoretical Relevance | Hypothesized Effect | Indicator & Coding Scheme |
|---|---|---|---|---|
| Ruminant contamination in stored water supply given uncontaminated source water | The extent of fecal contamination from ruminants (cattle, goats, and sheep) in the household’s main stored drinking water supply. | Outcome variable | Outcome variable | Indicator: Detection of BacR molecular marker in a 100mL sample of the household’s stored water Coding: 1 = BacR MST molecular marker detected, 0 = Otherwise |
| Proximity of ruminant feces production to stored water | Presence of ruminants and/or their feces within compound of household from which stored water sample was taken | Ruminant presence in the compound makes feces in the environment likely and be difficult to keep out of stored water supply due to proximity. | Positively associated with likelihood of contamination | Indicator: Self-reported number of ruminants living in the compound Coding: 1 = Some ruminants live in household compound 0 = No ruminants live in household compound |
| Opportunity for introduction of contamination into stored water | Length of time water is stored, which may allow ruminant fecal contamination to enter stored drinking water supply | The longer the time that water is in storage before use, the greater the chance that contaminated objects (e.g., hands, utensils etc.) will be introduced into the water | Positively associated with likelihood of contamination | Indicator: The number of hours that have elapsed since currently stored water was fetched from the source, as reported by the respondent. Coding *: 0.95 = Water self-reported as collected 1 day ago 0.5 = Water self-reported as collected 4.5 h ago 0.05 = Water self-reported as collected 1 h ago * These values were provided as calibration points for the fsQCA software to score a range of storage times from 1 h to 2 weeks. |
| Unsafe water extraction method | The extent to which a household’s method for extracting water from storage is likely to result in contamination of the water, e.g., by hands and other contaminated objects | Retrieving water using hands and other potentially contaminated objects allows for the (re)contamination of the stored water supply. | Positively associated with likelihood of contamination | Indicator: Observed method of water retrieval when respondent asked to get a cup of water for child (or themself, if no child) Coding: 1 = Respondent inserted hands or an object into the water; 0 = Respondent poured water from the top of the storage container |
| Lack of supportive household environment for handwashing | The extent to which household members lack regular access to supplies essential for good hand hygiene | The absence of soap is hypothesized to increase the likelihood of BacR contamination in the stored water supply | Positively associated with likelihood of contamination | Indicator: Observed presence of water and soap together at the home handwashing station 1: Water and soap not at home handwashing station when enumerators visited 0: Water and soap at home handwashing station when enumerators visited |
| Assay | Binary Analysis | Binary Metric | Quantitative Metric | |||
|---|---|---|---|---|---|---|
| Sensitivity | Specificity | Sensitivity | Specificity | Sensitivity | Specificity | |
| HumM2 | 100% | 60% | yes | no | no | no |
| HF183 | 100% | 48% | yes | no | no | yes |
| BacHum | 80% | 44% | yes | no | no | no |
| Rum2Bac | 100% | 67% | yes | no | yes | yes |
| BacR | 100% | 87% | yes | yes | yes | yes |
| BacCow | 100% | 60% | yes | no | yes | no |
| Avian GFD | 100% | 48% | yes | no | no | yes |
| Metric | Average (Range) | |
|---|---|---|
| Household (HH) size | 5 (2–10) | |
| Formal Education, mother | Primary education completed (none to post-secondary) | |
| Assets | Household has Electricity | 9% (4 HHs) |
| Bicycle Ownership | 42% (19HHs) | |
| Mobile Phone Ownership | 76% (34 HHs) | |
| Water Source | Spring | 84% (38 HHs) |
| Well | 11.1% (5 HHs) | |
| Stream | 2% (1 HH) | |
| Borehole | 2% (1 HH) | |
| Toilet | Access to toilet facility | 100% (45 HHs) |
| Private Use | 47% (21 HHs) | |
| For shared, mean number of households sharing | 3.2 HHs | |
| Animal Ownership | Cattle | 47% (21 HHs) |
| Goats | 4% (2 HHs) | |
| Sheep | 7% (3 HHs) | |
| Poultry | 76% (34 HHs) | |
| Dogs | 31% (14 HHs) | |
| Cats | 9% (4HHs) | |
| HH Water Treatment | 13% (6 HHs) |
| Assay | Both Source and Stored Water Contaminated | Both Source and Stored Water Uncontaminated | Only Source Water Contaminated | Only Stored Water Contaminated |
|---|---|---|---|---|
| BacR | 11 (24%) | 4 (9%) | 1 (2%) | 29 (64%) |
| HF183 | 1 (2%) | 42 (93%) | 2 (4%) | 0 (0%) |
| Avian GFD | 0 (0%) | 43 (96%) | 0 (0%) | 2 (4%) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hamzah, L.; Boehm, A.B.; Davis, J.; Pickering, A.J.; Wolfe, M.; Mureithi, M.; Harris, A. Ruminant Fecal Contamination of Drinking Water Introduced Post-Collection in Rural Kenyan Households. Int. J. Environ. Res. Public Health 2020, 17, 608. https://doi.org/10.3390/ijerph17020608
Hamzah L, Boehm AB, Davis J, Pickering AJ, Wolfe M, Mureithi M, Harris A. Ruminant Fecal Contamination of Drinking Water Introduced Post-Collection in Rural Kenyan Households. International Journal of Environmental Research and Public Health. 2020; 17(2):608. https://doi.org/10.3390/ijerph17020608
Chicago/Turabian StyleHamzah, Latifah, Alexandria B. Boehm, Jennifer Davis, Amy J. Pickering, Marlene Wolfe, Maryanne Mureithi, and Angela Harris. 2020. "Ruminant Fecal Contamination of Drinking Water Introduced Post-Collection in Rural Kenyan Households" International Journal of Environmental Research and Public Health 17, no. 2: 608. https://doi.org/10.3390/ijerph17020608
APA StyleHamzah, L., Boehm, A. B., Davis, J., Pickering, A. J., Wolfe, M., Mureithi, M., & Harris, A. (2020). Ruminant Fecal Contamination of Drinking Water Introduced Post-Collection in Rural Kenyan Households. International Journal of Environmental Research and Public Health, 17(2), 608. https://doi.org/10.3390/ijerph17020608