Enhanced Biogas Production by Ligninolytic Strain Enterobacter hormaechei KA3 for Anaerobic Digestion of Corn Straw
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation and Identification of the Ligninolytic Strain
2.2. Measurement of the Lignin-Degrading Ability of the Ligninolytic Strain
2.2.1. Determination of the Lignin Degradation Rate
2.2.2. Scanning Electron Microscope Analysis of the Pretreated CS
2.3. Effects of the Ligninolytic Strain on the AD of CS
2.3.1. Substrate and Inoculum
2.3.2. Experimental Design
2.3.3. Analytical Methods
2.4. DNA Extraction, PCR Amplification, and Illumina Sequencing
2.5. Statistical Analysis
3. Results and Discussion
3.1. Screening and Identification of Ligninolytic Strains
3.2. Lignin-Degrading Capability of Enterobacter Hormaechei KA3
3.2.1. Lignin-Degrading Rate
3.2.2. Enzyme Activity
3.2.3. Intuitive Analysis of Strain KA3 with CS Treatment
3.3. Effect of Strain KA3 on AD of CS
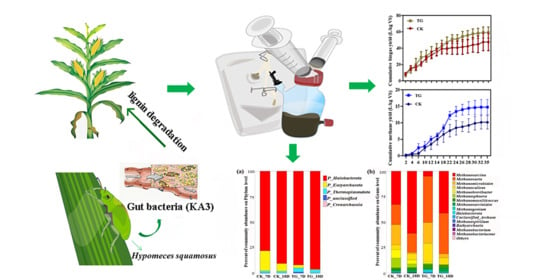
3.3.1. Biogas/Methane Production
3.3.2. Substrate Conversion Rate
3.4. Effects of Strain KA3 on the Microbial Community in the AD System
3.4.1. Archaeal Community Characteristics
3.4.2. Bacterial Community Characteristics
3.5. Practical Implementation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AD | anaerobic digestion |
| CS | corn straw |
| LB | Luria–Bertaini |
| SW | saline water |
| SEM | scanning electron microscope |
| CM | cow manure |
| TG | treated group |
| CK | control group |
| VFA | volatile fatty acid |
| COD | chemical oxygen demand |
| 7D | digested for seven days |
| 18D | digested for 18 days |
| 22D | digested for 22 days |
| TS | total solid |
| VS | volatile solid |
| TOC | total organic carbon |
| SD | standard deviation |
| LiP | lignin peroxidase |
| MnP | manganese peroxidase |
| Lac | laccase |
| ASOWs | agricultural solid organic waste |
References
- Pavlas, M.; Dvořáček, J.; Pitschke, T.; Peche, R. Biowaste Treatment and Waste-To-Energy—Environmental Benefits. Energies 2020, 13, 1994. [Google Scholar] [CrossRef] [Green Version]
- Mancini, G.; Papirio, S.; Lens, P.; Esposito, G. A Preliminary Study of the Effect of Bioavailable Fe and Co on the Anaerobic Digestion of Rice Straw. Energies 2019, 12, 577. [Google Scholar] [CrossRef] [Green Version]
- Li, P.; He, C.; Li, G.; Ding, P.; Lan, M.; Gao, Z.; Jiao, Y. Biological pretreatment of corn straw for enhancing degradation efficiency and biogas production. Bioengineered 2020, 11, 251–260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandes, T.V.; Bos, G.J.; Zeeman, G.; Sanders, J.P.; van Lier, J.B. Effects of thermo-chemical pre-treatment on anaerobic biodegradability and hydrolysis of lignocellulosic biomass. Bioresour. Technol. 2009, 100, 2575–2579. [Google Scholar] [CrossRef] [PubMed]
- Wagner, A.O.; Lackner, N.; Mutschlechner, M.; Prem, E.M.; Markt, R.; Illmer, P. Biological Pretreatment Strategies for Second-Generation Lignocellulosic Resources to Enhance Biogas Production. Energies 2018, 11, 1797. [Google Scholar] [CrossRef] [Green Version]
- Tsegaye, B.; Balomajumder, C.; Roy, P. Biodegradation of wheat straw by Ochrobactrum oryzae BMP03 and Bacillus sp. BMP01 bacteria to enhance biofuel production by increasing total reducing sugars yield. Environ. Sci. Pollut. Res. Int. 2018, 25, 30585–30596. [Google Scholar] [CrossRef]
- Xu, R.; Zhang, K.; Liu, P.; Han, H.; Zhao, S.; Kakade, A.; Khan, A.; Du, D.; Li, X. Lignin depolymerization and utilization by bacteria. Bioresour. Technol. 2018, 269, 557–566. [Google Scholar] [CrossRef] [PubMed]
- Arredondo-Santoyo, M.; Herrera-Camacho, J.; Vazquez-Garciduenas, M.S.; Vazquez-Marrufo, G. Corn stover induces extracellular laccase activity in Didymosphaeria sp. (syn. = Paraconiothyrium sp.) and exhibits increased in vitro ruminal digestibility when treated with this fungal species. Folia Microbiol. 2020, 65, 849–861. [Google Scholar] [CrossRef]
- Singh, S.; Harms, H.; Schlosser, D. Screening of ecologically diverse fungi for their potential to pretreat lignocellulosic bioenergy feedstock. Appl. Microbiol. Biotechnol. 2014, 98, 3355–3370. [Google Scholar] [CrossRef]
- Suman, S.K.; Dhawaria, M.; Tripathi, D.; Raturi, V.; Adhikari, D.K.; Kanaujia, P.K. Investigation of lignin biodegradation by Trabulsiella sp. isolated from termite gut. Int. Biodeterior. Biodegrad. 2016, 112, 12–17. [Google Scholar] [CrossRef]
- Zhou, H.; Guo, W.; Xu, B.; Teng, Z.; Tao, D.; Lou, Y.; Gao, Y. Screening and identification of lignin-degrading bacteria in termite gut and the construction of LiP-expressing recombinant Lactococcus lactis. Microb. Pathog. 2017, 112, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Wong, L.J.; H’ng, P.S.; Wong, S.Y.; Lee, S.H.; Lum, W.C.; Chai, E.W.; Wong, W.Z.; Chin, K.L. Termite digestomes as a potential source of symbiotic microbiota for lignocelluloses degradation: A review. Biol. Sci. 2014, 17, 956–963. [Google Scholar]
- Available online: https://baike.baidu.com/item/%E7%BB%BF%E7%A3%B7%E8%B1%A1%E7%94%B2/3486165?fromtitle=%E8%93%9D%E7%BB%BF%E8%B1%A1&fromid=7901507&fr=aladdin (accessed on 1 August 1997).
- Xu, F.F.; Chen, J.; Husson, J. Leaf area lost, rather than herbivore type, determines the induction of extrafloral nectar secretion in a tropical plant (Clerodendrum philippinum). Arthropod-Plant Interact. 2014, 8, 513–518. [Google Scholar] [CrossRef]
- Li, M.; Geng, Z.; Liao, P.; Wang, X.; Yang, T.; Wang, J.; Wang, D.; Gao, L.; Du, B. Trial marriage model-Female mate choice under male interference. J. Anim. Ecol. 2020, 89, 1851–1859. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Li, Q.; Lin, Y.; Lin, X.; Dai, Y.; Guo, Z.; Pan, D. Screening of a microbial consortium for selective degradation of lignin from tree trimmings. Bioresour. Technol. 2018, 254, 247–255. [Google Scholar] [CrossRef] [PubMed]
- Maganhotto de Souza Silva, C.M.; Soares de Melo, I.; Roberto de Oliveira, P. Ligninolytic enzyme production by Ganoderma spp. Enzym. Microb. Technol. 2005, 37, 324–329. [Google Scholar] [CrossRef]
- Nyári, J.; Kakuk, B.; Bagi, Z.; Rákhely, G.; Kovács, K.L. Use of ensiled green willow biomass in biogas fermentation. Biol. Futur. 2021. [Google Scholar] [CrossRef]
- Choi, G.-W.; Kang, H.-W.; Kim, Y.-R.; Chung, B.-W. Ethanol production by Zymomonas mobilis CHZ2501 from industrial starch feedstocks. Biotechnol. Bioprocess Eng. 2009, 13, 765–771. [Google Scholar] [CrossRef]
- Wang, X.; Dong, T.; Zhang, A.; Fang, Y.; Chen, D.; Zhao, C.; Luo, Q.; Yang, H. Isolation of bacteria capable of hydrogen production in dark fermentation and intensification of anaerobic granular sludge activity. Int. J. Hydrogen Energy 2019, 44, 15853–15862. [Google Scholar] [CrossRef]
- Zhao, J.; Jing, Y.; Zhang, J.; Sun, Y.; Wang, Y.; Wang, H.; Bi, X. Aged refuse enhances anaerobic fermentation of food waste to produce short-chain fatty acids. Bioresour. Technol. 2019, 289, 121547. [Google Scholar] [CrossRef]
- Yu, J.; Zhao, Y.; Liu, B.; Zhao, Y.; Wu, J.; Yuan, X.; Zhu, W.; Cui, Z. Accelerated acidification by inoculation with a microbial consortia in a complex open environment. Bioresour. Technol. 2016, 216, 294–301. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Zhang, J.; Lu, M.; Qin, C.; Chen, Y.; Yang, L.; Huang, Q.; Wang, J.; Shen, Z.; Shen, Q. Microbial communities of an arable soil treated for 8 years with organic and inorganic fertilizers. Biol. Fertil. Soils 2016, 52, 455–467. [Google Scholar] [CrossRef]
- Yang, C.X.; Wang, T.; Gao, L.N.; Yin, H.J.; Lu, X. Isolation, identification and characterization of lignin-degrading bacteria from Qinling, China. J. Appl. Microbiol. 2017, 123, 1447–1460. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Zhao, Y.; Meng, L.; Wang, X.; Yi, Y.; Shan, Y.; Liu, B.; Zhou, Y.; Lü, X. Isolation of Thermostable Lignocellulosic Bacteria From Chicken Manure Compost and a M42 Family Endocellulase Cloning From Geobacillus thermodenitrificans Y7. Front. Microbiol. 2020, 11, 281. [Google Scholar] [CrossRef] [PubMed]
- Barai, S.; Chattopadhyay, L.; Majumdar, B. Studies on delignification in jute (Corchorus spp L.) fibre with promising lignin degrading bacterial isolates. J. Environ. Biol. 2020, 41, 703–710. [Google Scholar] [CrossRef]
- Zhang, K.; Xu, R.; Abomohra, A.E.-F.; Xie, S.; Yu, Z.; Guo, Q.; Liu, P.; Peng, L.; Li, X. A sustainable approach for efficient conversion of lignin into biodiesel accompanied by biological pretreatment of corn straw. Energy Convers. Manag. 2019, 199, 111928. [Google Scholar] [CrossRef]
- Rahimi, H.; Sattler, M.L.; Hossain, M.D.S.; Rodrigues, J.L.M. Boosting landfill gas production from lignin-containing wastes via termite hindgut microorganism. Waste Manag. 2020, 105, 299–308. [Google Scholar] [CrossRef]
- Li, Y.; Zhao, J.; Achinas, S.; Zhang, Z.; Krooneman, J.; Euverink, G.J.W. The biomethanation of cow manure in a continuous anaerobic digester can be boosted via a bioaugmentation culture containing Bathyarchaeota. Sci. Total Environ. 2020, 745, 141042. [Google Scholar] [CrossRef]
- Shah, T.A.; Ali, S.; Afzal, A.; Tabassum, R. Simultaneous Pretreatment and Biohydrogen Production from Wheat Straw by Newly Isolated Ligninolytic Bacillus Sp. Strains with Two-Stage Batch Fermentation System. Bioenergy Res. 2018, 11, 835–849. [Google Scholar] [CrossRef]
- Baba, Y.; Matsuki, Y.; Takizawa, S.; Suyama, Y.; Tada, C.; Fukuda, Y.; Saito, M.; Nakai, Y. Pretreatment of Lignocellulosic Biomass with Cattle Rumen Fluid for Methane Production: Fate of Added Rumen Microbes and Indigenous Microbes of Methane Seed Sludge. Microbes Environ. 2019, 34, 421–428. [Google Scholar] [CrossRef] [Green Version]
- Azizi, A.; Sharifi, A.; Fazaeli, H.; Azarfar, A.; Jonker, A.; Kiani, A. Effect of transferring lignocellulose-degrading bacteria from termite to rumen fluid of sheep on in vitro gas production, fermentation parameters, microbial populations and enzyme activity. J. Integr. Agric. 2020, 19, 1323–1331. [Google Scholar] [CrossRef]
- Aguirre-Villegas, H.A.; Larson, R.A.; Sharara, M.A. Anaerobic digestion, solid-liquid separation, and drying of dairy manure: Measuring constituents and modeling emission. Sci. Total Environ. 2019, 696, 134059. [Google Scholar] [CrossRef]
- Musatti, A.; Ficara, E.; Mapelli, C.; Sambusiti, C.; Rollini, M. Use of solid digestate for lignocellulolytic enzymes production through submerged fungal fermentation. J. Environ. Manag. 2017, 199, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Boon, N.; Top, E.M.; Verstraete, W.; Siciliano, S.D. Bioaugmentation as a tool to protect the structure and function of an activated-sludge microbial community against a 3-chloroaniline shock load. Appl. Environ. Microbiol. 2003, 69, 1511–1520. [Google Scholar] [CrossRef] [Green Version]
- Zumstein, E.; Moletta, R. Examination of two years of community dynamics in ananaerobic bioreactor using uorescence polymerasechain reaction (PCR) single-strand conformationpolymorphism analysis. Environ. Microbiol. 2000, 2, 69–78. [Google Scholar] [CrossRef]
- Carballa, M.; Smits, M.; Etchebehere, C.; Boon, N.; Verstraete, W. Correlations between molecular and operational parameters in continuous lab-scale anaerobic reactors. Appl. Microbiol. Biotechnol. 2011, 89, 303. [Google Scholar] [CrossRef]
- Li, D.; Ni, H.; Jiao, S.; Lu, Y.; Zhou, J.; Sun, B.; Liang, Y. Coexistence patterns of soil methanogens are closely tied to methane generation and community assembly in rice paddies. Microbiome 2021, 9, 20. [Google Scholar] [CrossRef]
- Nelson, M.C.; Morrison, M.; Yu, Z. A meta-analysis of the microbial diversity observed in anaerobic digesters. Bioresour. Technol. 2011, 102, 3730–3739. [Google Scholar] [CrossRef]
- Schmidt, O.; Hink, L.; Horn, M.A.; Drake, H.L. Peat: Home to novel syntrophic species that feed acetate- and hydrogen-scavenging methanogens. ISME J. 2016, 10, 1954–1966. [Google Scholar] [CrossRef]
- Pytlak, A.; Kasprzycka, A. Szafranek-Nakonieczna A, Grzadziel J, Kubaczynski A, Proc K; et al. Biochar addition reinforces microbial interspecies cooperation in methanation of sugar beet waste (pulp). Sci. Total Environ. 2020, 730, 138921. [Google Scholar] [CrossRef]
- Ping, Q.; Lu, X.; Zheng, M.; Li, Y. Effect of CaO2 addition on anaerobic digestion of waste activated sludge at different temperatures and the promotion of valuable carbon source production under ambient condition. Bioresour. Technol. 2018, 265, 247–256. [Google Scholar] [CrossRef]
- Deng, Y.; Huang, Z.; Ruan, W.; Miao, H.; Shi, W.; Zhao, M. Enriching ruminal polysaccharide-degrading consortia via co-inoculation with methanogenic sludge and microbial mechanisms of acidification across lignocellulose loading gradients. Appl. Microbiol. Biotechnol. 2018, 102, 3819–3830. [Google Scholar] [CrossRef] [PubMed]
- Ruifang, G. Microbial diversity in a full-scale anaerobic reactor treating high concentration organic cassava wastewater. Afr. J. Biotechnol. 2012, 11, 6494–6500. [Google Scholar] [CrossRef]
- Guo, W.; Li, Y.; Zhao, K.; Xu, Q.; Jiang, H.; Zhou, H. Performance and Microbial Community Analysis of Anaerobic Digestion of Vinegar Residue with Adding of Acetylene Black or Hydrochar. Waste and Biomass Valorization 2019, 11, 3315–3325. [Google Scholar] [CrossRef]
- Jin, W.; Xu, X.; Yang, F. Application of Rumen Microorganisms for Enhancing Biogas Production of Corn Straw and Livestock Manure in a Pilot-Scale Anaerobic Digestion System: Performance and Microbial Community Analysis. Energies 2018, 11, 920. [Google Scholar] [CrossRef] [Green Version]
- David, A.; Govil, T.; Tripathi, A.; McGeary, J.; Farrar, K.; Sani, R. Thermophilic Anaerobic Digestion: Enhanced and Sustainable Methane Production from Co-Digestion of Food and Lignocellulosic Wastes. Energies 2018, 11, 2058. [Google Scholar] [CrossRef] [Green Version]





| Category | Samples | Shannon | Simpson | ACE | Chao1 |
|---|---|---|---|---|---|
| Archaea | CK-7D | 1.69 | 0.37 | 75.51 | 73.38 |
| CK-18D | 1.97 | 0.24 | 69.22 | 73.33 | |
| TG-7D | 2.17 | 0.22 | 131.78 | 96.75 | |
| TG-18D | 2.30 | 0.16 | 108.76 | 94.67 | |
| Bacteria | CK-7D | 3.91 | 0.06 | 667.44 | 654.36 |
| CK-18D | 3.76 | 0.06 | 693.33 | 701.43 | |
| TG-7D | 4.00 | 0.05 | 706.00 | 697.75 | |
| TG-18D | 4.20 | 0.03 | 713.06 | 705.25 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Q.; Zhang, J.; Zhao, S.; Song, P.; Chen, Y.; Liu, P.; Mao, C.; Li, X. Enhanced Biogas Production by Ligninolytic Strain Enterobacter hormaechei KA3 for Anaerobic Digestion of Corn Straw. Energies 2021, 14, 2990. https://doi.org/10.3390/en14112990
Zhang Q, Zhang J, Zhao S, Song P, Chen Y, Liu P, Mao C, Li X. Enhanced Biogas Production by Ligninolytic Strain Enterobacter hormaechei KA3 for Anaerobic Digestion of Corn Straw. Energies. 2021; 14(11):2990. https://doi.org/10.3390/en14112990
Chicago/Turabian StyleZhang, Qing, Jing Zhang, Shuai Zhao, Peizhi Song, Yanli Chen, Pu Liu, Chunlan Mao, and Xiangkai Li. 2021. "Enhanced Biogas Production by Ligninolytic Strain Enterobacter hormaechei KA3 for Anaerobic Digestion of Corn Straw" Energies 14, no. 11: 2990. https://doi.org/10.3390/en14112990
APA StyleZhang, Q., Zhang, J., Zhao, S., Song, P., Chen, Y., Liu, P., Mao, C., & Li, X. (2021). Enhanced Biogas Production by Ligninolytic Strain Enterobacter hormaechei KA3 for Anaerobic Digestion of Corn Straw. Energies, 14(11), 2990. https://doi.org/10.3390/en14112990