Targeted Polymer-Based Probes for Fluorescence Guided Visualization and Potential Surgery of EGFR-Positive Head-and-Neck Tumors
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Physico-Chemical Characterization
2.3. Synthesis of Monomers and Polymer Precursors
2.4. Synthesis of Peptide-Targeted Nanoprobes
2.5. Synthesis of hEGF-Targeted Nanoprobes
2.6. Synthesis of Monoclonal Antibody (mAb)-Targeted Nanoprobe
2.7. Synthesis of Star Polymers
2.8. Cell Culture
2.9. Flow Cytometry
2.10. Statistical Analysis
2.11. Immunofluorescence Analysis of Cells
2.12. Immunofluorescence Analysis of Tissues
2.13. Antibodies
2.14. Fluorescence Microscopy
2.15. Intravital Tumor Accumulation Assessment
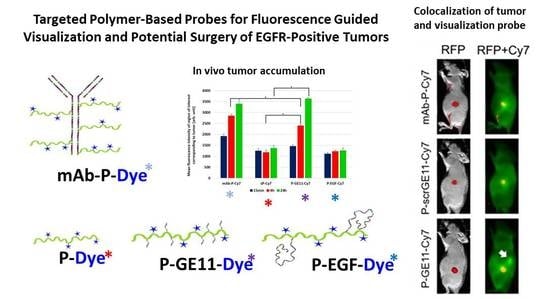
3. Results
3.1. Synthesis of the Polymer Nanoprobes
3.2. Binding Affinity of Polymeric Conjugates to EGFR-Positive Cells
3.3. EGFR Expression
3.4. Tumor Accumulation Based on Molecular Weight of Conjugates
3.5. Tumor Accumulation Based on Active Targeting
4. Discussion
4.1. Synthesis of the Polymer Nanoprobes
4.2. Binding Affinity of Polymeric Conjugates to EGFR-Positive Cells
4.3. Tumor Accumulation Based on Molecular Weight of Conjugates Versus Active Targeting
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Baddour, H.M. The Importance of Margins in Head and Neck Cancer. J. Surg. Oncol. 2016, 113, 248–255. [Google Scholar] [CrossRef] [PubMed]
- Kamat, M. A comprehensive review of surgical margin in oral squamous cell carcinoma highlighting the significance of tumor-free surgical margins. J. Cancer Res. Ther. 2019, 15, 449–454. [Google Scholar] [CrossRef] [PubMed]
- Zanoni, D.K.; Migliacci, J.C.; Xu, B.; Katabi, N.; Montero, P.H.; Ganly, I.; Shah, J.P.; Wong, R.J.; Ghossein, R.A.; Patel, S.G. A Proposal to Redefine Close Surgical Margins in Squamous Cell Carcinoma of the Oral Tongue. JAMA Otolaryngol. Neck Surg. 2017, 143, 555. [Google Scholar] [CrossRef]
- Dik, E.A.; Willems, S.M.; Ipenburg, N.A.; Adriaansens, S.O.; Rosenberg, A.J.W.P.; Van Es, R.J.J. Resection of early oral squamous cell carcinoma with positive or close margins: Relevance of adjuvant treatment in relation to local recurrence: Margins of 3 mm as safe as 5 mm. Oral Oncol. 2014. [Google Scholar] [CrossRef] [PubMed]
- Zabrodsky, M.; Lukes, P.; Lukesova, E.; Boucek, J.; Plzak, J. The Role of Narrow Band Imaging in the Detection of Recurrent Laryngeal and Hypopharyngeal Cancer after Curative Radiotherapy. Biomed Res. Int. 2014. [Google Scholar] [CrossRef] [PubMed]
- Miles, B.A.; Patsias, A.; Quang, T.; Polydorides, A.D.; Richards-Kortum, R.; Sikora, A.G. Operative margin control with high-resolution optical microendoscopy for head and neck squamous cell carcinoma. Laryngoscope 2015. [Google Scholar] [CrossRef]
- Liu, W.; Wang, Y.M.; Li, Y.H.; Cai, S.J.; Yin, X.B.; He, X.W.; Zhang, Y.K. Fluorescent Imaging-Guided Chemotherapy-and-Photodynamic Dual Therapy with Nanoscale Porphyrin Metal–Organic Framework. Small 2017. [Google Scholar] [CrossRef]
- Bethune, G.; Bethune, D.; Ridgway, N.; Xu, Z. Epidermal growth factor receptor (EGFR) in lung cancer: An overview and update. J. Thorac. Dis. 2010, 2, 48–51. [Google Scholar]
- Ali, R.; Wendt, M.K. The paradoxical functions of EGFR during breast cancer progression. Signal Transduct. Target. Ther. 2017, 2, 16042. [Google Scholar] [CrossRef]
- Walker, F.; Abramowitz, L.; Benabderrahmane, D.; Duval, X.; Descatoire, V.; Hénin, D.; Lehy, T.; Aparicio, T. Growth factor receptor expression in anal squamous lesions: Modifications associated with oncogenic human papillomavirus and human immunodeficiency virus. Hum. Pathol. 2009, 40, 1517–1527. [Google Scholar] [CrossRef]
- Xu, H.; Zong, H.; Ma, C.; Ming, X.; Shang, M.; Li, K.; He, X.; Du, H.; Cao, L. Epidermal growth factor receptor in glioblastoma. Oncol. Lett. 2017, 14, 512–516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zimmermann, M.; Zouhair, A.; Azria, D.; Ozsahin, M. The epidermal growth factor receptor (EGFR) in head and neck cancer: Its role and treatment implications. Radiat. Oncol. 2006, 1, 11. [Google Scholar] [CrossRef] [PubMed]
- Pola, R.; Parnica, J.; Zuska, K.; Bohmová, E.; Filipová, M.; Pechar, M.; Pankrác, J.; Mucksová, J.; Trefil, P.; Kalina, J.; et al. Oligopeptide-targeted polymer nanoprobes for fluorescence-guided endoscopic surgery. Multifunct. Mater. 2019, 2, 24004. [Google Scholar] [CrossRef]
- Franovic, A.; Gunaratnam, L.; Smith, K.; Robert, I.; Patten, D.; Lee, S. Translational up-regulation of the EGFR by tumor hypoxia provides a nonmutational explanation for its overexpression in human cancer. Proc. Natl. Acad. Sci. USA 2007, 104, 13092–13097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orcutt, K.P.; Parsons, A.D.; Sibenaller, Z.A.; Scarbrough, P.M.; Zhu, Y.; Sobhakumari, A.; Wilke, W.W.; Kalen, A.L.; Goswami, P.; Miller, F.J.; et al. Erlotinib-Mediated Inhibition of EGFR Signaling Induces Metabolic Oxidative Stress through NOX4. Cancer Res. 2011, 71, 3932–3940. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maeda, H. Macromolecular therapeutics in cancer treatment: The EPR effect and beyond. J. Control. Release 2012, 164, 138–144. [Google Scholar] [CrossRef] [PubMed]
- Maeda, H.; Matsumura, Y. Tumoritropic and Lymphotropic Principles of Macromolecular Drugs. Crit. Rev. Ther. Drug Carrier Syst. 1989, 6, 193–210. [Google Scholar]
- Etrych, T.; Mrkvan, T.; Říhová, B.; Ulbrich, K. Star-shaped immunoglobulin-containing HPMA-based conjugates with doxorubicin for cancer therapy. J. Control. Release 2007, 122, 31–38. [Google Scholar] [CrossRef]
- Etrych, T.; Strohalm, J.; Kovář, L.; Kabešová, M.; Říhová, B.; Ulbrich, K. HPMA copolymer conjugates with reduced anti-CD20 antibody for cell-specific drug targeting. I. Synthesis and in vitro evaluation of binding efficacy and cytostatic activity. J. Control. Release 2009, 140, 18–26. [Google Scholar] [CrossRef]
- Etrych, T.; Strohalm, J.; Chytil, P.; Černoch, P.; Starovoytova, L.; Pechar, M.; Ulbrich, K. Biodegradable star HPMA polymer conjugates of doxorubicin for passive tumor targeting. Eur. J. Pharm. Sci. 2011, 42, 527–539. [Google Scholar] [CrossRef]
- Imagej. Available online: https://imagej.net (accessed on 11 October 2018).
- Li, S.; Schmitz, K.R.; Jeffrey, P.D.; Wiltzius, J.J.W.; Kussie, P.; Ferguson, K.M. Structural basis for inhibition of the epidermal growth factor receptor by cetuximab. Cancer Cell 2005, 7, 301–311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Genta, I.; Chiesa, E.; Colzani, B.; Modena, T.; Conti, B.; Dorati, R. GE11 peptide as an active targeting agent in antitumor therapy: A minireview. Pharmaceutics 2018, 10, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mazzuca, C.; Di Napoli, B.; Biscaglia, F.; Ripani, G.; Rajendran, S.; Braga, A.; Benna, C.; Mocellin, S.; Gobbo, M.; Meneghetti, M.; et al. Understanding the good and poor cell targeting activity of gold nanostructures functionalized with molecular units for the epidermal growth factor receptor. Nanoscale Adv. 2019, 1, 1970–1979. [Google Scholar] [CrossRef] [Green Version]
- Etrych, T.; Subr, V.; Strohalm, J.; Sírová, M.; Ríhová, B.; Ulbrich, K. HPMA copolymer-doxorubicin conjugates: The effects of molecular weight and architecture on biodistribution and in vivo activity. J. Control. Release 2012, 164, 346–354. [Google Scholar] [CrossRef] [PubMed]
- Seymour, L.W.; Miyamoto, Y.; Maeda, H.; Brereton, M.; Strohalm, J.; Ulbrich, K.; Duncan, R. Influence of molecular weight on passive tumour accumulation of a soluble macromolecular drug carrier. Eur. J. Cancer 1995, 31, 766–770. [Google Scholar] [CrossRef]
- Tolmachev, V.; Orlova, A.; Wei, Q.; Bruskin, A.; Carlsson, J.; Gedda, L. Comparative Biodistribution of Potential Anti- Glioblastoma Conjugates [ 111 In]DTPA-hEGF and [ 111 In]Bz-DTPA-hEGF in Normal Mice. Cancer Biother. Radiopharm. 2004, 19, 491–502. [Google Scholar] [CrossRef]
- Huang, L.; Gainkam, L.O.T.; Caveliers, V.; Vanhove, C.; Keyaerts, M.; De Baetselier, P.; Bossuyt, A.; Revets, H.; Lahoutte, T. SPECT Imaging with 99mTc-Labeled EGFR-Specific Nanobody for In Vivo Monitoring of EGFR Expression. Mol. Imaging Biol. 2008, 10, 167–175. [Google Scholar] [CrossRef]
- Liu, F.; Jiao, Y.; Jiao, Y.; Garcia-Godoy, F.; Gu, W.; Liu, Q. Sex difference in EGFR pathways in mouse kidney-potential impact on the immune system. BMC Genet. 2016, 17, 146. [Google Scholar] [CrossRef] [Green Version]









| Polymer Precursors and Conjugatesj | Mn | Mw/Mn | TTa or Hydrazide (mol%)/mAb or Peptide (wt%) | Mn,Td | Mn/Mn,Te | Dye (wt%) |
|---|---|---|---|---|---|---|
| P-TT | 36,800 | 1.14 | 8.8a | - | - | - |
| sP-TT | 15,300 | 1.89 | 5.9f | 11,800 | 1.31 | - |
| sP-MI | 19,000 | 1.94 | 5.9f | 15,800 | 1.18 | - |
| sP-Cy7 | 16,700 | 1.80 | - | - | - | 2.40c |
| sP-Dy-633-MI | 20,500 | 1.95 | - | 17,500 | 1.17 | 1.12b |
| sP-Cy7-MI | 20,500 | 1.95 | - | 18,200 | 1.13 | 1.30c |
| St-P-1 | 105,600 | 1.61 | 5.2 | - | - | - |
| St-P-2 | 445,000 | 1.73 | 5.2 | - | - | - |
| P-Dy-633 | 36,800 | 1.14 | - | - | - | 0.80b |
| P-Cy7 | 36,800 | 1.14 | - | - | - | 0.80c |
| P-GE11-Dy-633 | 39,500 | 1.14 | 15.0i | - | - | 0.80b |
| P-GE11-Cy7 | 39,500 | 1.14 | 15.0i | - | - | 0.76c |
| P-scrGE11-Cy7 | 39,500 | 1.14 | 15.0i | - | - | 0.83c |
| P-EGF-Dy-633 | 57,000 | 1.28 | 33.3g | - | - | 0.64b |
| P-EGF-Cy7 | 57,000 | 1.28 | 33.3g | - | - | 0.63c |
| mAb-P-Dy-633 | 262,500 | 1.60 | 45.2h | - | - | 0.50b |
| mAb-P-Cy7 | 262,500 | 1.60 | 44.0h | - | - | 0.62c |
| St-P-1-Cy7 | 109,000 | 1.65 | - | - | - | 2.43c |
| St-P-2-Cy7 | 457,000 | 1.75 | - | - | - | 2.40c |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pola, R.; Böhmová, E.; Filipová, M.; Pechar, M.; Pankrác, J.; Větvička, D.; Olejár, T.; Kabešová, M.; Poučková, P.; Šefc, L.; et al. Targeted Polymer-Based Probes for Fluorescence Guided Visualization and Potential Surgery of EGFR-Positive Head-and-Neck Tumors. Pharmaceutics 2020, 12, 31. https://doi.org/10.3390/pharmaceutics12010031
Pola R, Böhmová E, Filipová M, Pechar M, Pankrác J, Větvička D, Olejár T, Kabešová M, Poučková P, Šefc L, et al. Targeted Polymer-Based Probes for Fluorescence Guided Visualization and Potential Surgery of EGFR-Positive Head-and-Neck Tumors. Pharmaceutics. 2020; 12(1):31. https://doi.org/10.3390/pharmaceutics12010031
Chicago/Turabian StylePola, Robert, Eliška Böhmová, Marcela Filipová, Michal Pechar, Jan Pankrác, David Větvička, Tomáš Olejár, Martina Kabešová, Pavla Poučková, Luděk Šefc, and et al. 2020. "Targeted Polymer-Based Probes for Fluorescence Guided Visualization and Potential Surgery of EGFR-Positive Head-and-Neck Tumors" Pharmaceutics 12, no. 1: 31. https://doi.org/10.3390/pharmaceutics12010031
APA StylePola, R., Böhmová, E., Filipová, M., Pechar, M., Pankrác, J., Větvička, D., Olejár, T., Kabešová, M., Poučková, P., Šefc, L., Zábrodský, M., Janoušková, O., Bouček, J., & Etrych, T. (2020). Targeted Polymer-Based Probes for Fluorescence Guided Visualization and Potential Surgery of EGFR-Positive Head-and-Neck Tumors. Pharmaceutics, 12(1), 31. https://doi.org/10.3390/pharmaceutics12010031