The Biological Potential Hidden in Inclusion Bodies
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacteria Strains and Plasmids
2.2. Protein Production in L. lactis
2.3. Production and Purification of Soluble MMP-9
2.4. Purification of MMP-9 and mutMMP-9 IBs
2.5. Polymeric Micelles (PM) Synthesis
2.6. Transmission Electron Microscopy (TEM) Imaging
2.7. MMP-9 Loading Efficacy
2.8. X-ray Photoelectron Spectroscopy (XPS)
2.9. Protein Stability Assay
2.10. In Vivo Comparison of the Inflammatory Response to Solubilized MMP-9 versus MMP-9 IBs, Using a Mouse Model
2.11. Statistical Analysis
3. Results
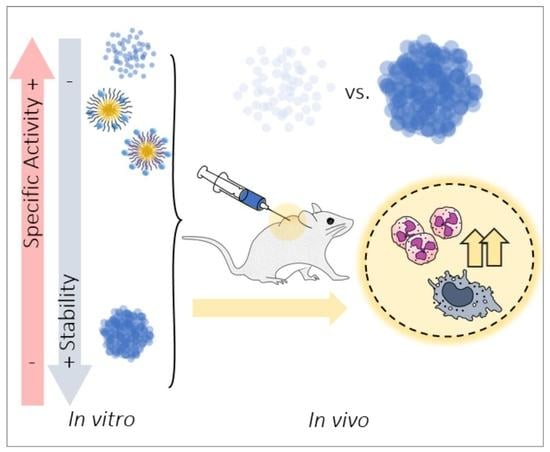
3.1. Activity of MMP-9 Nanoparticles
3.2. In Vitro MMP-9 Stability
3.3. In Vivo MMP-9 Activity
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- de Marco, A.; Ferrer-Miralles, N.; Garcia-Fruitós, E.; Mitraki, A.; Peternel, S.; Rinas, U.; Trujillo-Roldán, M.A.; Valdez-Cruz, N.A.; Vázquez, E.; Villaverde, A. Bacterial inclusion bodies are industrially exploitable amyloids. FEMS Microbiol. Rev. 2019, 43, 53–72. [Google Scholar] [CrossRef]
- García-Fruitós, E.; Vázquez, E.; Díez-Gil, C.; Corchero, J.L.; Seras-Franzoso, J.; Ratera, I.; Veciana, J.; Villaverde, A. Bacterial inclusion bodies: Making gold from waste. Trends Biotechnol. 2012, 30, 65–70. [Google Scholar] [CrossRef]
- García-Fruitós, E.; Seras-Franzoso, J.; Vazquez, E.; Villaverde, A. Tunable geometry of bacterial inclusion bodies as substrate materials for tissue engineering. Nanotechnology 2010, 21, 205101. [Google Scholar] [CrossRef] [PubMed]
- Seras-Franzoso, J.; Peebo, K.; Corchero, J.L.; Unzueta, U.; Rinas, U.; Dalby, M.J.; Vazquez, E.; García-fruitós, E.; Villaverde, A. A nanostructured bacterial bioscaffold for the sustained bottom-up delivery of protein drugs. Nanomedicine 2013, 8, 1587–1599. [Google Scholar] [CrossRef] [PubMed]
- Seras-Franzoso, J.; Peebo, K.; García-Fruitós, E.; Vázquez, E.; Rinas, U.; Villaverde, A. Improving protein delivery of fibroblast growth factor-2 from bacterial inclusion bodies used as cell culture substrates. Acta Biomater. 2014, 10, 1354–1359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seras-Franzoso, J.; Tatkiewicz, W.I.; Vazquez, E.; García-Fruitós, E.; Ratera, I.; Veciana, J.; Villaverde, A. Integrating mechanical and biological control of cell proliferation through bioinspired multieffector materials. Nanomedicine 2015, 10, 873–891. [Google Scholar] [CrossRef] [Green Version]
- Vázquez, E.; Corchero, J.L.; Burgueño, J.F.; Seras-Franzoso, J.; Kosoy, A.; Bosser, R.; Mendoza, R.; Martínez-Láinez, J.M.; Rinas, U.; Fernández, E.; et al. Functional inclusion bodies produced in bacteria as naturally occurring nanopills for advanced cell therapies. Adv. Mater. 2012, 24, 1742–1747. [Google Scholar] [CrossRef]
- Villaverde, A.; García-Fruitós, E.; Rinas, U.; Seras-Franzoso, J.; Kosoy, A.; Corchero, J.L.; Vazquez, E. Packaging protein drugs as bacterial inclusion bodies for therapeutic applications. Microb. Cell Fact. 2012, 11, 76. [Google Scholar] [CrossRef] [Green Version]
- Unzueta, U.; Cespedes, M.V.; Sala, R.; Alamo, P.; Sánchez-Chardi, A.; Pesarrodona, M.; Sánchez-García, L.; Cano-Garrido, O.; Villaverde, A.; Vázquez, E.; et al. Release of targeted protein nanoparticles from functional bacterial amyloids: A death star-like approach. J. Control. Release 2018, 279, 29–39. [Google Scholar] [CrossRef]
- Pesarrodona, M.; Jauset, T.; Díaz-Riascos, Z.V.; Sánchez-Chardi, A.; Beaulieu, M.; Seras-Franzoso, J.; Sánchez-García, L.; Baltà-Foix, R.; Mancilla, S.; Fernández, Y.; et al. Targeting Antitumoral Proteins to Breast Cancer by Local Administration of Functional Inclusion Bodies. Adv. Sci. 2019, 1900849. [Google Scholar] [CrossRef]
- Torrealba, D.; Parra, D.; Seras-Franzoso, J.; Vallejos-Vidal, E.; Yero, D.; Gibert, I.; Villaverde, A.; Garcia-Fruitós, E.; Roher, N. Nanostructured recombinant cytokines: A highly stable alternative to short-lived prophylactics. Biomaterials 2016, 107, 102–114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gifre-Renom, L.; Cano-Garrido, O.; Fàbregas, F.; Roca-Pinilla, R.; Seras-Franzoso, J.; Ferrer-Miralles, N.; Villaverde, A.; Bach, A.; Devant, M.; Arís, A.; et al. A new approach to obtain pure and active proteins from Lactococcus lactis protein aggregates. Sci. Rep. 2018, 8, 13917. [Google Scholar] [CrossRef] [PubMed]
- Villaverde, A.; Corchero, J.L.; Seras-Franzoso, J.; Garcia-Fruitós, E. Functional protein aggregates: Just the tip of the iceberg. Nanomedicine 2015, 10, 2881–2891. [Google Scholar] [CrossRef] [PubMed]
- Vandooren, J.; Van Den Steen, P.E.; Opdenakker, G. Biochemistry and molecular biology of gelatinase B or matrix metalloproteinase-9 (MMP-9): The next decade. Crit. Rev. Biochem. Mol. Biol. 2013, 48, 222–272. [Google Scholar] [CrossRef]
- Kolaczkowska, E.; Chadzinska, M.; Scislowska-Czarnecka, A.; Plytycz, B.; Opdenakker, G.; Arnold, B. Gelatinase B/matrix metalloproteinase-9 contributes to cellular infiltration in a murine model of zymosan peritonitis. Immunobiology 2006, 211, 137–148. [Google Scholar] [CrossRef]
- Cortes-Perez, N.G.; Poquet, I.; Oliveira, M.; Gratadoux, J.J.; Madsen, S.M.; Miyoshi, A.; Corthier, G.; Azevedo, V.; Langella, P.; Bermúdez-Humarán, L.G. Construction and characterization of a Lactococcus lactis strain deficient in intracellular ClpP and extracellular HtrA proteases. Microbiology 2006, 152, 2611–2618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poquet, I.; Saint, V.; Seznec, E.; Simoes, N.; Bolotin, A.; Gruss, A. HtrA is the unique surface housekeeping protease in Lactococcus lactis and is required for natural protein processing. Mol. Microbiol. 2000, 35, 1042–1051. [Google Scholar] [CrossRef]
- Cano-Garrido, O.; Sánchez-Chardi, A.; Parés, S.; Giró, I.; Tatkiewicz, W.I.; Ferrer-Miralles, N.; Ratera, I.; Natalello, A.; Cubarsi, R.; Veciana, J.; et al. Functional protein-based nanomaterial produced in GRAS microorganism: A new platform for biotechnology. Acta Biomater. 2016, 43, 230–239. [Google Scholar] [CrossRef] [Green Version]
- Rowsell, S.; Hawtin, P.; Minshull, C.A.; Jepson, H.; Brockbank, S.M.V.; Barratt, D.G.; Slater, A.M.; McPheat, W.L.; Waterson, D.; Henney, A.M.; et al. Crystal structure of human MMP9 in complex with a reverse hydroxamate inhibitor. J. Mol. Biol. 2002, 319, 173–181. [Google Scholar] [CrossRef]
- Roderfeld, M.; Weiskirchen, R.; Wagner, S.; Berres, M.; Henkel, C.; Gro, J.; Gressner, A.M.; Matern, S.; Roeb, E. Inhibition of hepatic fibrogenesis by matrix metallo- proteinase-9 mutants in mice. FASEB J. 2006, 20, 444–454. [Google Scholar] [CrossRef]
- Rafael, D.; Martínez, F.; Andrade, F.; Seras-Franzoso, J.; Garcia-Aranda, N.; Gener, P.; Sayós, J.; Arango, D.; Abasolo, I.; Schwartz, S., Jr. Efficient EFGR mediated siRNA delivery to breast cancer cells by Cetuximab functionalized Pluronic® F127/Gelatin. Chem. Eng. J. 2018, 340, 81–93. [Google Scholar] [CrossRef]
- Dubois, B.; Masure, S.; Hurtenbach, U.; Paemen, L.; Heremans, H.; Oord, J.; Van Den Sciot, R.; Meinhardt, T.; Hämmerling, G.; Opdenakker, G.; et al. Resistance of young gelatinase B—Deficient mice to experimental autoimmune encephalomyelitis and necrotizing tail lesions. J. Clin. Investig. 1999, 104, 1507–1515. [Google Scholar] [CrossRef] [PubMed]
- Vandooren, J.; Berghmans, N.; Dillen, C.; Van Aelst, I.; Ronsse, I.; Israel, L.L.; Rosenberger, I.; Kreuter, J.; Lellouche, J.-P.; Michaeli, S.; et al. Intradermal air pouch leukocytosis as an in vivo test for nanoparticles. Int. J. Nanomed. 2013, 8, 4745–4756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Céspedes, M.V.; Cano-Garrido, O.; Álamo, P.; Sala, R.; Gallardo, A.; Serna, N.; Falgàs, A.; Voltà-Durán, E.; Casanova, I.; Sánchez-Chardi, A.; et al. Engineering secretory amyloids for remote and highly selective destruction of metastatic foci. Adv Mater. 2019, 1907348. [Google Scholar] [CrossRef] [PubMed]
- Rinas, U.; Garcia-Fruitós, E.; Corchero, J.L.; Vázquez, E.; Seras-Franzoso, J.; Villaverde, A. Bacterial inclusion bodies: Discovering their better half. Trends Biochem. Sci. 2017, 42, 726–737. [Google Scholar] [CrossRef] [PubMed]




© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gifre-Renom, L.; Seras-Franzoso, J.; Rafael, D.; Andrade, F.; Cano-Garrido, O.; Martinez-Trucharte, F.; Ugarte-Berzal, E.; Martens, E.; Boon, L.; Villaverde, A.; et al. The Biological Potential Hidden in Inclusion Bodies. Pharmaceutics 2020, 12, 157. https://doi.org/10.3390/pharmaceutics12020157
Gifre-Renom L, Seras-Franzoso J, Rafael D, Andrade F, Cano-Garrido O, Martinez-Trucharte F, Ugarte-Berzal E, Martens E, Boon L, Villaverde A, et al. The Biological Potential Hidden in Inclusion Bodies. Pharmaceutics. 2020; 12(2):157. https://doi.org/10.3390/pharmaceutics12020157
Chicago/Turabian StyleGifre-Renom, Laia, Joaquin Seras-Franzoso, Diana Rafael, Fernanda Andrade, Olivia Cano-Garrido, Francesc Martinez-Trucharte, Estefania Ugarte-Berzal, Erik Martens, Lise Boon, Antonio Villaverde, and et al. 2020. "The Biological Potential Hidden in Inclusion Bodies" Pharmaceutics 12, no. 2: 157. https://doi.org/10.3390/pharmaceutics12020157
APA StyleGifre-Renom, L., Seras-Franzoso, J., Rafael, D., Andrade, F., Cano-Garrido, O., Martinez-Trucharte, F., Ugarte-Berzal, E., Martens, E., Boon, L., Villaverde, A., Opdenakker, G., Schwartz, S., Jr., Arís, A., & Garcia-Fruitós, E. (2020). The Biological Potential Hidden in Inclusion Bodies. Pharmaceutics, 12(2), 157. https://doi.org/10.3390/pharmaceutics12020157