Multi-Organs-on-Chips for Testing Small-Molecule Drugs: Challenges and Perspectives
Abstract
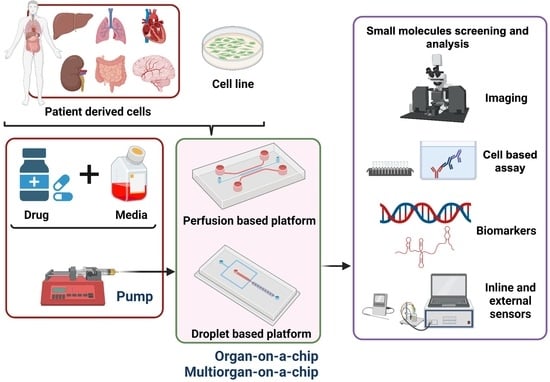
:1. Introduction
2. Small Molecules: Properties and Applications
3. Organs-on-Chips
3.1. Fabrication Methods
3.2. Drug Assays
4. High-Throughput Applications and Current Challenges
4.1. Fabrication Challenges
4.2. High-Throughput Challenges
4.2.1. Liver Platforms
4.2.2. Lung Platforms
4.3. Tumor Platforms
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Herland, A.; Maoz, B.M.; Das, D.; Somayaji, M.R.; Prantil-Baun, R.; Novak, R.; Cronce, M.; Huffstater, T.; Jeanty, S.S.F.; Ingram, M.; et al. Quantitative prediction of human pharmacokinetic responses to drugs via fluidically coupled vascularized organ chips. Nat. Biomed. Eng. 2020, 4, 421–436. [Google Scholar] [CrossRef] [PubMed]
- Kimura, H.; Sakai, Y.; Fujii, T. Organ/body-on-a-chip based on microfluidic technology for drug discovery. Drug Metab. Pharmacokinet. 2018, 33, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Bailey, J.; Thew, M.; Balls, M. An Analysis of the Use of Animal Models in Predicting Human Toxicology and Drug Safety. Altern. Lab. Anim. 2014, 42, 181–199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kingwell, K. 3D cell technologies head to the R&D assembly line. Nat. Rev. Drug Discov. 2017, 16, 6–7. [Google Scholar] [CrossRef]
- Langer, R.; Vacanti, J.P. Tissue engineering. Science 1993, 260, 920–926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barros, A.S.; Costa, A.; Sarmento, B. Building three-dimensional lung models for studying pharmacokinetics of inhaled drugs. Adv. Drug Deliv. Rev. 2021, 170, 386–395. [Google Scholar] [CrossRef] [PubMed]
- Skardal, A.; Shupe, T.; Atala, A. Organoid-on-a-chip and body-on-a-chip systems for drug screening and disease modeling. Drug Discov. Today 2016, 21, 1399–1411. [Google Scholar] [CrossRef]
- Park, D.; Lee, J.; Chung, J.J.; Jung, Y.; Kim, S.H. Integrating Organs-on-Chips: Multiplexing, Scaling, Vascularization, and Innervation. Trends Biotechnol. 2020, 38, 99–112. [Google Scholar] [CrossRef]
- Jalili-Firoozinezhad, S.; Miranda, C.C.; Cabral, J.M. Modeling the Human Body on Microfluidic Chips. Trends Biotechnol. 2021, 39, 838–852. [Google Scholar] [CrossRef]
- Maschmeyer, I.; Lorenz, A.K.; Schimek, K.; Hasenberg, T.; Ramme, A.P.; Hübner, J.; Lindner, M.; Drewell, C.; Bauer, S.; Thomas, A.; et al. A four-organ-chip for interconnected long-term co-culture of human intestine, liver, skin and kidney equivalents. Lab Chip 2015, 15, 2688–2699. [Google Scholar] [CrossRef] [Green Version]
- De Mello, C.P.P.; Carmona-Moran, C.A.; McAleer, C.W.; Perez, J.; Coln, E.A.; Long, C.J.; Oleaga, C.; Riu, A.; Note, R.; Teissier, S.; et al. Microphysiological heart–liver body-on-a-chip system with a skin mimic for evaluating topical drug delivery. Lab Chip 2020, 20, 749–759. [Google Scholar] [CrossRef] [PubMed]
- Sasserath, T.; Rumsey, J.W.; McAleer, C.W.; Bridges, L.R.; Long, C.J.; Elbrecht, D.; Schuler, F.; Roth, A.; Bertinetti-LaPatki, C.; Shuler, M.L.; et al. Differential Monocyte Actuation in a Three-Organ Functional Innate Immune System-on-a-Chip. Adv. Sci. 2020, 7, 2000323. [Google Scholar] [CrossRef]
- Villenave, R.; Wales, S.Q.; Hamkins-Indik, T.; Papafragkou, E.; Weaver, J.C.; Ferrante, T.C.; Bahinski, A.; Elkins, C.; Kulka, M.; Ingber, D.E. Human Gut-On-A-Chip Supports Polarized Infection of Coxsackie B1 Virus In Vitro. PLoS ONE 2017, 12, e0169412. [Google Scholar] [CrossRef] [PubMed]
- Cifuente, J.; Ferrer, M.F.; de Giusti, C.J.; Song, W.-C.; Romanowski, V.; Hafenstein, S.L.; Gómez, R.M. Molecular determinants of disease in coxsackievirus B1 murine infection. J. Med. Virol. 2011, 83, 1571–1581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, Z.; Guo, Z.; Yang, C.-T.; Prestidge, C.; Thierry, B. “Mucus-on-Chip”: A new tool to study the dynamic penetration of nanoparticulate drug carriers into mucus. Int. J. Pharm. 2021, 598, 120391. [Google Scholar] [CrossRef]
- Kamaly, N.; Yameen, B.; Wu, J.; Farokhzad, O.C. Degradable Controlled-Release Polymers and Polymeric Nanoparticles: Mechanisms of Controlling Drug Release. Chem. Rev. 2016, 116, 2602–2663. [Google Scholar] [CrossRef] [Green Version]
- Sebe, I.; Szabó, P.; Kállai-Szabó, B.; Zelkó, R. Incorporating small molecules or biologics into nanofibers for optimized drug release: A review. Int. J. Pharm. 2015, 494, 516–530. [Google Scholar] [CrossRef]
- Zhang, Y.; Fang, C.; Carvalho, W.S.P.; Gao, Y.; Serpe, M.J. Triggered Small-Molecule Release from Dual-Stimuli Responsive Microgels. ACS Appl. Polym. Mater. 2021, 3, 410–417. [Google Scholar] [CrossRef]
- Hill, L.K.; Meleties, M.; Katyal, P.; Xie, X.; Delgado-Fukushima, E.G.; Jihad, T.; Liu, C.-F.; O’Neill, S.C.; Tu, R.S.; Renfrew, P.D.; et al. Thermoresponsive Protein-Engineered Coiled-Coil Hydrogel for Sustained Small Molecule Release. Biomacromolecules 2019, 20, 3340–3351. [Google Scholar] [CrossRef]
- Morihiro, K.; Ankenbruck, N.; Lukasak, B.; Deiters, A. Small Molecule Release and Activation through DNA Computing. J. Am. Chem. Soc. 2017, 139, 13909–13915. [Google Scholar] [CrossRef]
- Santini, J.T.; Cima, M.J.; Langer, R. A controlled-release microchip. Nat. Cell Biol. 1999, 397, 335–338. [Google Scholar] [CrossRef]
- Zhang, B.; Korolj, A.; Lai, B.F.L.; Radisic, M. Advances in organ-on-a-chip engineering. Nat. Rev. Mater. 2018, 3, 257–278. [Google Scholar] [CrossRef]
- Huh, D.; Matthews, B.D.; Mammoto, A.; Montoya-Zavala, M.; Hsin, H.Y.; Ingber, D.E. Reconstituting Organ-Level Lung Functions on a Chip. Science 2010, 328, 1662–1668. [Google Scholar] [CrossRef] [Green Version]
- Ferrari, E.; Rasponi, M. Liver–Heart on chip models for drug safety. APL Bioeng. 2021, 5, 031505. [Google Scholar] [CrossRef]
- Li, X.-G.; Chen, M.-X.; Zhao, S.-Q.; Wang, X.-Q. Intestinal Models for Personalized Medicine: From Conventional Models to Microfluidic Primary Intestine-on-a-chip. Stem Cell Rev. Rep. 2021, 1–15. [Google Scholar] [CrossRef]
- Sun, A.M.; Hoffman, T.; Luu, B.Q.; Ashammakhi, N.; Li, S. Application of lung microphysiological systems to COVID-19 modeling and drug discovery: A review. Bio-Des. Manuf. 2021, 1–19. [Google Scholar] [CrossRef]
- Phillips, J.A.; Grandhi, T.S.P.; Davis, M.; Gautier, J.-C.; Hariparsad, N.; Keller, D.; Sura, R.; Van Vleet, T.R. A pharmaceutical industry perspective on microphysiological kidney systems for evaluation of safety for new therapies. Lab Chip 2020, 20, 468–476. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Oleaga, C.; Long, C.J.; Esch, M.B.; McAleer, C.W.; Miller, P.G.; Hickman, J.J.; Shuler, M.L. Self-contained, low-cost Body-on-a-Chip systems for drug development. Exp. Biol. Med. 2017, 242, 1701–1713. [Google Scholar] [CrossRef] [PubMed]
- Kolahchi, A.R.; Mohtaram, N.K.; Modarres, H.P.; Mohammadi, M.H.; Geraili, A.; Jafari, P.; Akbari, M.; Sanati-Nezhad, A. Microfluidic-Based Multi-Organ Platforms for Drug Discovery. Micromachines 2016, 7, 162. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Kankala, R.K.; Wang, S.-B.; Chen, A.-Z. Multi-Organs-on-Chips: Towards Long-Term Biomedical Investigations. Molecules 2019, 24, 675. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.B.; Sung, J.H. Organ-on-a-chip technology and microfluidic whole-body models for pharmacokinetic drug toxicity screening. Biotechnol. J. 2013, 8, 1258–1266. [Google Scholar] [CrossRef]
- Van der Zanden, S.Y.; Luimstra, J.; Neefjes, J.; Borst, J.; Ovaa, H. Opportunities for Small Molecules in Cancer Immunotherapy. Trends Immunol. 2020, 41, 493–511. [Google Scholar] [CrossRef] [PubMed]
- Makurvet, F.D. Biologics vs. small molecules: Drug costs and patient access. Med. Drug Discov. 2021, 9, 100075. [Google Scholar] [CrossRef]
- Ngo, H.X.; Garneau-Tsodikova, S. What are the drugs of the future? MedChemComm 2018, 9, 757–758. [Google Scholar] [CrossRef] [PubMed]
- Warner, K.D.; Hajdin, C.E.; Weeks, K.M. Principles for targeting RNA with drug-like small molecules. Nat. Rev. Drug Discov. 2018, 17, 547–558. [Google Scholar] [CrossRef]
- Govardhanagiri, S.; Bethi, S.; Nagaraju, G.P. Small-Molecules and Pancreatic Cancer Trials and Troubles. In Breaking Tolerance to Pancreatic Cancer Unresponsiveness to Chemotherapy; Academic Press: Cambridge, MA, USA, 2019. [Google Scholar] [CrossRef]
- Ma, C.; Zhao, L.; Zhou, E.-M.; Xu, J.; Shen, S.; Wang, J. On-Chip Construction of Liver Lobule-like Microtissue and Its Application for Adverse Drug Reaction Assay. Anal. Chem. 2016, 88, 1719–1727. [Google Scholar] [CrossRef] [PubMed]
- Wagner, I.; Materne, E.-M.; Brincker, S.; Süssbier, U.; Frädrich, C.; Busek, M.; Sonntag, F.; Sakharov, D.A.; Trushkin, E.V.; Tonevitsky, A.; et al. A dynamic multi-organ-chip for long-term cultivation and substance testing proven by 3D human liver and skin tissue co-culture. Lab Chip 2013, 13, 3538–3547. [Google Scholar] [CrossRef] [Green Version]
- Edington, C.D.; Chen, W.L.K.; Geishecker, E.; Kassis, T.; Soenksen, L.R.; Bhushan, B.M.; Freake, D.; Kirschner, J.; Maass, C.; Tsamandouras, N.; et al. Interconnected Microphysiological Systems for Quantitative Biology and Pharmacology Studies. Sci. Rep. 2018, 8, 4530. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Zhang, Y.S.; Zhang, X.; Liu, C. Organ-on-a-chip platforms for accelerating the evaluation of nanomedicine. Bioact. Mater. 2021, 6, 1012–1027. [Google Scholar] [CrossRef]
- Miller, P.G.; Shuler, M.L. Design and demonstration of a pumpless 14 compartment microphysiological system. Biotechnol. Bioeng. 2016, 113, 2213–2227. [Google Scholar] [CrossRef]
- Tang, Y.; Soroush, F.; Sheffield, J.B.; Wang, B.; Prabhakarpandian, B.; Kiani, M.F. A Biomimetic Microfluidic Tumor Microenvironment Platform Mimicking the EPR Effect for Rapid Screening of Drug Delivery Systems. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef]
- Park, J.Y.; Jang, J.; Kang, H.-W. 3D Bioprinting and its application to organ-on-a-chip. Microelectron. Eng. 2018, 200, 1–11. [Google Scholar] [CrossRef]
- Lee, H.; Cho, D.-W. One-step fabrication of an organ-on-a-chip with spatial heterogeneity using a 3D bioprinting technology. Lab Chip 2016, 16, 2618–2625. [Google Scholar] [CrossRef] [Green Version]
- Convery, N.; Gadegaard, N. 30 years of microfluidics. Micro Nano Eng. 2019, 2, 76–91. [Google Scholar] [CrossRef]
- Spivey, E.C.; Xhemalce, B.; Shear, J.B.; Finkelstein, I.J. 3D-Printed Microfluidic Microdissector for High-Throughput Studies of Cellular Aging. Anal. Chem. 2014, 86, 7406–7412. [Google Scholar] [CrossRef] [Green Version]
- Nakao, Y.; Kimura, H.; Sakai, Y.; Fujii, T. Bile canaliculi formation by aligning rat primary hepatocytes in a microfluidic device. Biomicrofluidics 2011, 5, 022212. [Google Scholar] [CrossRef] [Green Version]
- Sung, J.H.; Yu, J.; Luo, D.; Shuler, M.L.; March, J.C. Microscale 3-D hydrogel scaffold for biomimetic gastrointestinal (GI) tract model. Lab Chip 2011, 11, 389–392. [Google Scholar] [CrossRef]
- Grafton, M.M.; Wang, L.; Vidi, P.-A.; Leary, J.; Lelievre, S. Breast on-a-chip: Mimicry of the channeling system of the breast for development of theranostics. Integr. Biol. 2011, 3, 451–459. [Google Scholar] [CrossRef]
- Au, S.; Chamberlain, M.; Mahesh, S.; Sefton, M.V.; Wheeler, A.R. Hepatic organoids for microfluidic drug screening. Lab Chip 2014, 14, 3290–3299. [Google Scholar] [CrossRef] [PubMed]
- Grover, W.; von Muhlen, M.; Manalis, S.R. Teflon films for chemically-inert microfluidic valves and pumps. Lab Chip 2008, 8, 913–918. [Google Scholar] [CrossRef] [PubMed]
- Chudobova, D.; Cihalova, K.; Skalickova, S.; Zitka, J.; Rodrigo, M.A.M.; Milosavljevic, V.; Hynek, D.; Kopel, P.; Vesely, R.; Adam, V.; et al. 3D-printed chip for detection of methicillin-resistant Staphylococcus aureuslabeled with gold nanoparticles. Electrophoresis 2015, 36, 457–466. [Google Scholar] [CrossRef]
- Krejcova, L.; Nejdl, L.; Rodrigo, M.A.M.; Zurek, M.; Matousek, M.; Hynek, D.; Zitka, O.; Kopel, P.; Adam, V.; Kizek, R. 3D printed chip for electrochemical detection of influenza virus labeled with CdS quantum dots. Biosens. Bioelectron. 2014, 54, 421–427. [Google Scholar] [CrossRef]
- Domansky, K.; Sliz, J.D.; Wen, N.; Hinojosa, C.; Thompson, G.; Fraser, J.P.; Hamkins-Indik, T.; Hamilton, G.A.; Levner, D.; Ingber, D.E. SEBS elastomers for fabrication of microfluidic devices with reduced drug absorption by injection molding and extrusion. Microfluid. Nanofluidics 2017, 21, 107. [Google Scholar] [CrossRef]
- Yi, H.; Wu, L.-Q.; Ghodssi, R.; Rubloff, G.; Payne, G.F.; Bentley, W.E. Signal-Directed Sequential Assembly of Biomolecules on Patterned Surfaces. Langmuir 2005, 21, 2104–2107. [Google Scholar] [CrossRef]
- Yi, H.; Wu, L.Q.; Bentley, W.E.; Ghodssi, R.; Rubloff, G.W.; Culver, J.N.; Payne, G.F. Biofabrication with Chitosan. Biomacromolecules 2005, 6, 2881–2894. [Google Scholar] [CrossRef]
- Bettinger, C.J.; Cyr, K.M.; Matsumoto, A.; Langer, R.; Borenstein, J.T.; Kaplan, D.L. Silk Fibroin Microfluidic Devices. Adv. Mater. 2007, 19, 2847–2850. [Google Scholar] [CrossRef]
- Pérez-Rigueiro, J.; Viney, C.; Llorca, J.; Elices, M. Mechanical properties of single-brin silkworm silk. J. Appl. Polym. Sci. 2000, 75, 1270–1277. [Google Scholar] [CrossRef]
- Schimek, K.; Busek, M.; Brincker, S.; Groth, B.; Hoffmann, S.; Lauster, R.; Lindner, G.; Lorenz, A.; Menzel, U.; Sonntag, F.; et al. Integrating biological vasculature into a multi-organ-chip microsystem. Lab Chip 2013, 13, 3588–3598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hasenberg, T.; Mühleder, S.; Dotzler, A.; Bauer, S.; Labuda, K.; Holnthoner, W.; Redl, H.; Lauster, R.; Marx, U. Emulating human microcapillaries in a multi-organ-chip platform. J. Biotechnol. 2015, 216, 1–10. [Google Scholar] [CrossRef]
- Ling, Y.; Rubin, J.; Deng, Y.; Huang, C.; Demirci, U.; Karp, J.M.; Khademhosseini, A. A cell-laden microfluidic hydrogel. Lab Chip 2007, 7, 756–762. [Google Scholar] [CrossRef] [PubMed]
- Rahfoth, B.; Weisser, J.; Sternkopf, F.; Aigner, T.; Von Der Mark, K.; Bräuer, R. Transplantation of allograft chondrocytes embedded in agarose gel into cartilage defects of rabbits. Osteoarthr. Cartil. 1998, 6, 50–65. [Google Scholar] [CrossRef] [Green Version]
- Sugioka, K.; Hanada, Y.; Midorikawa, K. 3D microstructuring of glass by femtosecond laser direct writing and application to biophotonic microchips. Prog. Electromagn. Res. Lett. 2008, 1, 181–188. [Google Scholar] [CrossRef] [Green Version]
- Hanada, Y.; Sugioka, K.; Shihira-Ishikawa, I.; Kawano, H.; Miyawaki, A.; Midorikawa, K. 3D microfluidic chips with integrated functional microelements fabricated by a femtosecond laser for studying the gliding mechanism of cyanobacteria. Lab Chip 2011, 11, 2109–2115. [Google Scholar] [CrossRef]
- Sosa-Hernández, J.E.; Villalba-Rodríguez, A.M.; Romero-Castillo, K.D.; Aguilar-Aguila-Isaías, M.A.; García-Reyes, I.E. Organs-on-a-chip module: A review from the development and applications perspective. Micromachines 2018, 9, 536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, T.F.; Siow, K.S.; Ng, P.Y.; Majlis, B.Y. Enhancing the biocompatibility of the polyurethane methacrylate and off-stoichiometry thiol-ene polymers by argon and nitrogen plasma treatment. Mater. Sci. Eng. C 2017, 79, 613–621. [Google Scholar] [CrossRef] [PubMed]
- Kuo, J.S.; Ng, L.; Yen, G.S.; Lorenz, R.M.; Schiro, P.G.; Edgar, J.S.; Zhao, Y.; Lim, D.S.W.; Allen, P.B.; Jeffries, G.D.M.; et al. A new USP Class VI-compliant substrate for manufacturing disposable microfluidic devices. Lab Chip 2009, 9, 870–876. [Google Scholar] [CrossRef] [Green Version]
- Cherpinski, A.; Torres-Giner, S.; Vartiainen, J.; Peresin, M.S.; Lahtinen, P.; Lagaron, J.M. Improving the water resistance of nanocellulose-based films with polyhydroxyalkanoates processed by the electrospinning coating technique. Cellulose 2018, 25, 1291–1307. [Google Scholar] [CrossRef]
- Alfadhel, A.; Quyang, J.; Mahajan, C.G.; Forouzandeh, F.; Cormier, D.; Borkholder, D.A. Inkjet printed polyethylene glycol as a fugitive ink for the fabrication of flexible microfluidic systems. Mater. Design 2018, 150, 182–187. [Google Scholar] [CrossRef] [PubMed]
- Shim, K.; Kim, S.H.; Lee, N.; Kim, B.; Kim, T.H.; Jung, Y.; Choi, N.; Sung, J.H. Fabrication of micrometer-scale porous gelatin scaffolds for 3D cell culture. J. Ind. Eng. Chem. 2017, 50, 183–189. [Google Scholar] [CrossRef]
- Zamboni, F.; Keays, M.; Hayes, S.; Albadarin, A.; Walker, G.; Kiely, P.A.; Collins, M.N. Enhanced cell viability in hyaluronic acid coated poly(lactic-co-glycolic acid) porous scaffolds within microfluidic channels. Int. J. Pharm. 2017, 532, 595–602. [Google Scholar] [CrossRef]
- Miller, J.; Stevens, K.R.; Yang, M.T.; Baker, B.; Nguyen, D.-H.T.; Cohen, D.; Toro, E.; Chen, A.A.; Galie, P.; Yu, X.; et al. Rapid casting of patterned vascular networks for perfusable engineered three-dimensional tissues. Nat. Mater. 2012, 11, 768–774. [Google Scholar] [CrossRef] [PubMed]
- Bertassoni, L.E.; Cecconi, M.; Manoharan, V.; Nikkhah, M.; Hjortnaes, J.; Cristino, A.L.; Barabaschi, G.; Demarchi, D.; Dokmeci, M.R.; Yang, Y.; et al. Hydrogel bioprinted microchannel networks for vascularization of tissue engineering constructs. Lab Chip 2014, 14, 2202–2211. [Google Scholar] [CrossRef] [Green Version]
- Porter, S. Human immune response to recombinant human proteins. J. Pharm. Sci. 2011, 90, 1–11. [Google Scholar] [CrossRef]
- Perini, P.; Facchinetti, A.; Bulian, P.; Massaro, A.R.; De Pascalis, D.; Bertolotto, A.; Biasi, G.; Gallo, P. Interferon-beta (INF-b) antibodies in interferon-b1a-and interferon-b1b-treated multiple sclerosis patients. Prevalence, kinetics, cross-reactivity, and factors enhancing interferon-b immunogenicity in vivo. Eur. Cytokine Netw. 2001, 12, 56–61. [Google Scholar] [PubMed]
- Casadevall, N.; Nataf, J.; Viron, B.; Kolta, A.; Kiladjian, J.-J.; Martin-Dupont, P.; Michaud, P.; Papo, T.; Ugo, V.; Teyssandier, I.; et al. Pure Red-Cell Aplasia and Antierythropoietin Antibodies in Patients Treated with Recombinant Erythropoietin. N. Engl. J. Med. 2002, 346, 469–475. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, S.; Barger, T.; Zhou, L.; Hale, M.; Mytych, D.; Gupta, S.; Swanson, S.J.; Civoli, F. Strategy to confirm the presence of anti-erythropoietin neutralizing antibodies in human serum. J. Pharm. Biomed. Anal. 2011, 55, 1265–1274. [Google Scholar] [CrossRef] [PubMed]
- Sung, J.H.; Shuler, M.L. A micro cell culture analog (µCCA) with 3-D hydrogel culture of multiple cell lines to assess metabolism-dependent cytotoxicity of anti-cancer drugs. Lab Chip 2009, 9, 1385–1394. [Google Scholar] [CrossRef]
- Skardal, A.; Murphy, S.V.; Devarasetty, M.; Mead, I.; Kang, H.-W.; Seol, Y.-J.; Zhang, Y.S.; Shin, S.-R.; Zhao, L.; Aleman, J.; et al. Multi-tissue interactions in an integrated three-tissue organ-on-a-chip platform. Sci. Rep. 2017, 7, 1–16. [Google Scholar] [CrossRef]
- Maharjan, S.; Cecen, B.; Zhang, Y.S. 3D Immunocompetent Organ-on-a-Chip Models. Small Methods 2020, 4, 2000235. [Google Scholar] [CrossRef]
- Qureshi, Z.P.; Seoane-Vazquez, E.; Rodriguez-Monguio, R.; Stevenson, K.B.; Szeinbach, S.L. Market withdrawal of new molecular entities approved in the United States from 1980 to 2009. Pharmacoepidemiol. Drug Saf. 2011, 20, 772–777. [Google Scholar] [CrossRef]
- Kang, Y.B.; Rawat, S.; Duchemin, N.; Bouchard, M.; Noh, M. Human Liver Sinusoid on a Chip for Hepatitis B Virus Replication Study. Micromachines 2017, 8, 27. [Google Scholar] [CrossRef] [Green Version]
- Si, L.; Bai, H.; Rodas, M.; Cao, W.; Oh, C.Y.; Jiang, A.; Nurani, A.; Zhu, D.Y.; Goyal, G.; Gilpin, S.E.; et al. Human organs-on-chips as tools for repurposing approved drugs as potential influenza and COVID19 therapeutics in viral pandemics. BioRxiv 2020. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020, 181, 271–280. [Google Scholar] [CrossRef]
- Paul, S.M.; Mytelka, D.; Dunwiddie, C.T.; Persinger, C.C.; Munos, B.H.; Lindborg, S.R.; Schacht, A.L. How to improve R&D productivity: The pharmaceutical industry’s grand challenge. Nat. Rev. Drug Discov. 2010, 9, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Pammolli, F.; Magazzini, L.; Riccaboni, M. The productivity crisis in pharmaceutical R&D. Nat. Rev. Drug Discov. 2011, 10, 428–438. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Doberstein, S.K. HTS technologies in biopharmaceutical discovery. Drug Discov. Today 2006, 11, 718–724. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.-H.; Huang, S.-B.; Lee, G.-B. Microfluidic cell culture systems for drug research. Lab Chip 2010, 10, 939–956. [Google Scholar] [CrossRef]
- Bhadriraju, K.; Chen, C.S. Engineering cellular microenvironments to improve cell-based drug testing. Drug Discov. Today 2002, 7, 612–620. [Google Scholar] [CrossRef]
- Damiati, S.; Kompella, U.B.; Damiati, S.A.; Kodzius, R. Microfluidic Devices for Drug Delivery Systems and Drug Screening. Genes 2018, 9, 103. [Google Scholar] [CrossRef] [Green Version]
- Miri, A.K.; Mostafavi, E.; Khorsandi, D.; Hu, S.-K.; Malpica, M.; Khademhosseini, A. Bioprinters for organs-on-chips. Biofabrication 2019, 11, 042002. [Google Scholar] [CrossRef]
- Sontheimer-Phelps, A.; Hassell, B.A.; Ingber, D.E. Modelling cancer in microfluidic human organs-on-chips. Nat. Rev. Cancer 2019, 19, 65–81. [Google Scholar] [CrossRef]
- Samavedi, S.; Joy, N. 3D printing for the development of in vitro cancer models. Curr. Opin. Biomed. Eng. 2017, 2, 35–42. [Google Scholar] [CrossRef]
- Ribas, J.; Sadeghi, H.; Manbachi, A.; Leijten, J.C.H.; Brinegar, K.; Zhang, Y.S.; Ferreira, L.; Khademhosseini, A. Cardiovascular Organ-on-a-Chip Platforms for Drug Discovery and Development. Appl. Vitr. Toxicol. 2016, 2, 82–96. [Google Scholar] [CrossRef] [Green Version]
- Van der Meer, A.D.; Poot, A.A.; Duits, M.H.G.; Feijen, J.; Vermes, I. Microfluidic Technology in Vascular Research. J. Biomed. Biotechnol. 2009, 2009, 1–10. [Google Scholar] [CrossRef]
- Gold, K.; Gaharwar, A.K.; Jain, A. Emerging trends in multiscale modeling of vascular pathophysiology: Organ-on-a-chip and 3D printing. Biomaterials 2019, 196, 2–17. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.; Yildirimer, L.; Khademhosseini, A.; Seifalian, A. Nanostructured Materials for Cardiovascular Tissue Engineering. J. Nanosci. Nanotechnol. 2012, 12, 4775–4785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tandon, N.; Cannizzaro, C.; Chao, P.-H.G.; Maidhof, R.; Marsano, A.; Au, H.T.H.; Radisic, M.; Vunjak-Novakovic, G. Electrical stimulation systems for cardiac tissue engineering. Nat. Protoc. 2009, 4, 155–173. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.S.; Aleman, J.; Arneri, A.; Bersini, S.; Piraino, F.; Shin, S.R.; Dokmeci, M.R.; Khademhosseini, A. From cardiac tissue engineering to heart-on-a-chip: Beating challenges. Biomed. Mater. 2015, 10, 034006. [Google Scholar] [CrossRef] [PubMed]
- Halldorsson, S.; Lucumi, E.; Gomez-Sjoberg, R.; Fleming, R.M. Advantages and challenges of microfluidic cell culture in polydimethylsiloxane devices. Biosens. Bioelectron. 2015, 63, 218–231. [Google Scholar] [CrossRef] [Green Version]
- Melin, J.; Quake, S.R. Microfluidic Large-Scale Integration: The Evolution of Design Rules for Biological Automation. Annu. Rev. Biophys. Biomol. Struct. 2007, 36, 213–231. [Google Scholar] [CrossRef] [Green Version]
- Hong, J.; Edel, J.B.; Demello, A.J. Micro-and nanofluidic systems for high-throughput biological screening. Drug Discov. Today 2009, 14, 134–146. [Google Scholar] [CrossRef]
- Wang, Z.; Kim, M.-C.; Marquez, M.; Thorsen, T. High-density microfluidic arrays for cell cytotoxicity analysis. Lab Chip 2007, 7, 740–745. [Google Scholar] [CrossRef] [PubMed]
- Dressler, O.J.; Maceiczyk, R.M.; Chang, S.I.; Demello, A.J. Droplet-Based Microfluidics: Enabling Impact on Drug Discovery. J. Biomol. Screen. 2013, 19, 483–496. [Google Scholar] [CrossRef] [PubMed]
- Beebe, D.J.; Mensing, G.A.; Walker, G.M. Physics and Applications of Microfluidics in Biology. Annu. Rev. Biomed. Eng. 2002, 4, 261–286. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.; Zhang, F.; Gao, M.; Niu, Y. Concentration Gradient Constructions Using Inertial Microfluidics for Studying Tumor Cell–Drug Interactions. Micromachines 2020, 11, 493. [Google Scholar] [CrossRef] [PubMed]
- Shourabi, A.Y.; Kashaninejad, N.; Saidi, M.S. An integrated microfluidic concentration gradient generator for mechanical stimulation and drug delivery. J. Sci. Adv. Mater. Devices 2021, 6, 280–290. [Google Scholar] [CrossRef]
- Coskun, A.F.; Su, T.-W.; Ozcan, A. Wide field-of-view lens-free fluorescent imaging on a chip. Lab Chip 2010, 10, 824–827. [Google Scholar] [CrossRef] [Green Version]
- Kemna, E.W.M.; Segerink, L.I.; Wolbers, F.; Vermes, I.; Berg, A.V.D. Label-free, high-throughput, electrical detection of cells in droplets. Analyst 2013, 138, 4585–4592. [Google Scholar] [CrossRef]
- Ferstl, W.; Klahn, T.; Schweikert, W.; Billeb, G.; Schwarzer, M.; Loebbecke, S. Inline Analysis in Microreaction Technology: A Suitable Tool for Process Screening and Optimization. Chem. Eng. Technol. 2007, 30, 370–378. [Google Scholar] [CrossRef]
- Sun, S.; Kennedy, R.T. Droplet Electrospray Ionization Mass Spectrometry for High Throughput Screening for Enzyme Inhibitors. Anal. Chem. 2014, 86, 9309–9314. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Q.; Kim, T. Review of microfluidic approaches for surface-enhanced Raman scattering. Sens. Actuators B Chem. 2016, 227, 504–514. [Google Scholar] [CrossRef]
- Polidoro, M.A.; Ferrari, E.; Marzorati, S.; Lleo, A.; Rasponi, M. Experimental liver models: From cell culture techniques to microfluidic organs-on-chip. Liver. Int. 2021, 41, 1744–1761. [Google Scholar] [CrossRef]
- Jang, K.-J.; Otieno, M.A.; Ronxhi, J.; Lim, H.-K.; Ewart, L.; Kodella, K.R.; Petropolis, D.B.; Kulkarni, G.; Rubins, J.E.; Conegliano, D.; et al. Reproducing human and cross-species drug toxicities using a Liver-Chip. Sci. Transl. Med. 2019, 11, eaax5516. [Google Scholar] [CrossRef]
- Moya, A.; Ortega-Ribera, M.; Guimerà, X.; Sowade, E.; Zea, M.; Illa, X.; Ramon, E.; Villa, R.; Gracia-Sancho, J.; Gabriel, G. Online oxygen monitoring using integrated inkjet-printed sensors in a liver-on-a-chip system. Lab Chip 2018, 18, 2023–2035. [Google Scholar] [CrossRef] [Green Version]
- Ma, L.-D.; Wang, Y.-T.; Wang, J.-R.; Wu, J.-L.; Meng, X.-S.; Hu, P.; Mu, X.; Liang, Q.-L.; Luo, G.-A. Design and fabrication of a liver-on-a-chip platform for convenient, highly efficient, and safe in situ perfusion culture of 3D hepatic spheroids. Lab Chip 2018, 18, 2547–2562. [Google Scholar] [CrossRef] [PubMed]
- Banaeiyan, A.A.; Theobald, J.; Paukštyte, J.; Wölfl, S.; Adiels, C.B.; Goksör, M. Design and fabrication of a scalable liver-lobule-on-a-chip microphysiological platform. Biofabrication 2017, 9, 15014. [Google Scholar] [CrossRef] [PubMed]
- Du, G.; Fang, Q.; den Toonder, J.M. Microfluidics for cell-based high throughput screening platforms—A review. Anal. Chim. Acta 2016, 903, 36–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- An, X.; Zuo, P.; Ye, B.-C. A single cell droplet microfluidic system for quantitative determination of food-borne pathogens. Talanta 2020, 209, 120571. [Google Scholar] [CrossRef] [PubMed]
- Occhetta, P.; Centola, M.; Tonnarelli, B.; Redaelli, A.; Martin, I.; Rasponi, M. High-throughput microfluidic platform for 3D cultures of mesenchymal stem cells. In 3D Cell Culture; Humana Press: New York, NY, USA, 2017; pp. 303–323. [Google Scholar]
- Fernandes, T.G.; Diogo, M.M.; Clark, D.S.; Dordick, J.S.; Cabral, J.M. High-throughput cellular microarray platforms: Applications in drug discovery, toxicology and stem cell research. Trends Biotechnol. 2009, 27, 342–349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeon, J.S.; Bersini, S.; Gilardi, M.; Dubini, G.; Charest, J.L.; Moretti, M.; Kamm, R.D. Human 3D vascularized organotypic microfluidic assays to study breast cancer cell extravasation. Proc. Natl. Acad. Sci. USA 2015, 112, E818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinton, D.A.; Daley, G.Q. The promise of induced pluripotent stem cells in research and therapy. Nat. Cell Biol. 2012, 481, 295–305. [Google Scholar] [CrossRef]
- Scott, C.W.; Peters, M.F.; Dragan, Y.P. Human induced pluripotent stem cells and their use in drug discovery for toxicity testing. Toxicol. Lett. 2013, 219, 49–58. [Google Scholar] [CrossRef]
- Deng, J.; Wei, W.; Chen, Z.; Lin, B.; Zhao, W.; Luo, Y.; Zhang, X. Engineered Liver-on-a-Chip Platform to Mimic Liver Functions and Its Biomedical Applications: A Review. Micromachines 2019, 10, 676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esch, M.B.; Mahler, G.J.; Stokol, T.; Shuler, M.L. Body-on-a-chip simulation with gastrointestinal tract and liver tissues suggests that ingested nanoparticles have the potential to cause liver injury. Lab Chip 2014, 14, 3081–3092. [Google Scholar] [CrossRef] [PubMed]
- Potkay, J.A. The promise of microfluidic artificial lungs. Lab Chip 2014, 14, 4122–4138. [Google Scholar] [CrossRef] [PubMed]
- Benam, K.H.; Villenave, R.; Lucchesi, C.; Varone, A.; Hubeau, C.; Lee, H.-H.; Alves, S.E.; Salmon, M.; Ferrante, T.C.; Weaver, J.C.; et al. Small airway-on-a-chip enables analysis of human lung inflammation and drug responses in vitro. Nat. Methods 2016, 13, 151–157. [Google Scholar] [CrossRef]
- Yang, X.; Li, K.; Zhang, X.; Liu, C.; Guo, B.; Wen, W.; Gao, X. Nanofiber membrane supported lung-on-a-chip microdevice for anti-cancer drug testing. Lab Chip 2018, 18, 486–495. [Google Scholar] [CrossRef]
- Li, W.; Sun, X.; Ji, B.; Yang, X.; Zhou, B.; Lu, Z.; Gao, X. PLGA Nanofiber/PDMS Microporous Composite Membrane-Sandwiched Microchip for Drug Testing. Micromachines 2020, 11, 1054. [Google Scholar] [CrossRef]
- Liu, Y.Y.; Sakolish, C.; Chen, Z.; Phan, D.T.; Bender, R.H.F.; Hughes, C.C.; Rusyn, I. Human in vitro vascularized micro-organ and micro-tumor models are reproducible organ-on-a-chip platforms for studies of anticancer drugs. Toxicology 2020, 445, 152601. [Google Scholar] [CrossRef]
- Bai, J.; Tu, T.-Y.; Kim, C.; Thiery, J.P.; Kamm, R.D. Identification of drugs as single agents or in combination to prevent carcinoma dissemination in a microfluidic 3D environment. Oncotarget 2015, 6, 36603–36614. [Google Scholar] [CrossRef] [Green Version]
- Lin, D.; Li, P.; Lin, J.; Shu, B.; Wang, W.; Zhang, Q.; Yang, N.; Liu, D.; Xu, B. Orthogonal Screening of Anticancer Drugs Using an Open-Access Microfluidic Tissue Array System. Anal. Chem. 2017, 89, 11976–11984. [Google Scholar] [CrossRef]
- Kamb, A. What’s wrong with our cancer models? Nat. Rev. Drug Discov. 2005, 4, 161–165. [Google Scholar] [CrossRef]
- Balkwill, F.R.; Capasso, M.; Hagemann, T. The tumor microenvironment at a glance. J. Cell Sci. 2012, 125, 5591–5596. [Google Scholar] [CrossRef] [Green Version]
- Gengenbacher, N.; Singhal, M.; Augustin, H.G. Preclinical mouse solid tumour models: Status quo, challenges and perspectives. Nat. Rev. Cancer 2017, 17, 751–765. [Google Scholar] [CrossRef]
- Joffe, A.R.; Bara, M.; Anton, N.; Nobis, N. The ethics of animal research: A survey of the public and scientists in North America. BMC Med. Ethic 2016, 17, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Kenney, R.M.; Loeser, A.; Whitman, N.A.; Lockett, M.R. Paper-based Transwell assays: An inexpensive alternative to study cellular invasion. Analyst 2019, 144, 206–211. [Google Scholar] [CrossRef]
- Huang, B.-W.; Gao, J.-Q. Application of 3D cultured multicellular spheroid tumor models in tumor-targeted drug delivery system research. J. Control. Release 2018, 270, 246–259. [Google Scholar] [CrossRef]
- Edmondson, R.; Broglie, J.J.; Adcock, A.F.; Yang, L. Three-Dimensional Cell Culture Systems and Their Applications in Drug Discovery and Cell-Based Biosensors. ASSAY Drug Dev. Technol. 2014, 12, 207–218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Del Piccolo, N.; Shirure, V.S.; Bi, Y.; Goedegebuure, S.P.; Gholami, S.; Hughes, C.C.; Fields, R.C.; George, S.C. Tumor-on-chip modeling of organ-specific cancer and metastasis. Adv. Drug Deliv. Rev. 2021, 175, 113798. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Fang, J.; Huang, S.; Wu, X.; Xie, X.; Wang, J.; Liu, F.; Zhang, M.; Peng, Z.; Hu, N. Tumor-on-a-chip: From bioinspired design to biomedical application. Microsyst. Nanoeng. 2021, 7, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Witkowski, M.T.; Harris, J.; Dolgalev, I.; Sreeram, S.; Qian, W.; Tong, J.; Chen, X.; Aifantis, I.; Chen, W. Leukemia-on-a-chip: Dissecting the chemoresistance mechanisms in B cell acute lymphoblastic leukemia bone marrow niche. Sci. Adv. 2020, 6, eaba5536. [Google Scholar] [CrossRef]
- Nguyen, D.-H.T.; Lee, E.; Alimperti, S.; Norgard, R.J.; Wong, A.; Lee, J.J.-K.; Eyckmans, J.; Stanger, B.Z.; Chen, C.S. A biomimetic pancreatic cancer on-chip reveals endothelial ablation via ALK7 signaling. Sci. Adv. 2019, 5, eaav6789. [Google Scholar] [CrossRef] [Green Version]
- Marturano-Kruik, A.; Nava, M.; Yeager, K.; Chramiec, A.; Hao, L.; Robinson, S.; Guo, E.; Raimondi, M.T.; Vunjak-Novakovic, G. Human bone perivascular niche-on-a-chip for studying metastatic colonization. Proc. Natl. Acad. Sci. USA 2018, 115, 1256–1261. [Google Scholar] [CrossRef] [Green Version]
- Ma, C.; Peng, Y.; Li, H.; Chen, W. Organ-on-a-Chip: A New Paradigm for Drug Development. Trends Pharmacol. Sci. 2021, 42, 119–133. [Google Scholar] [CrossRef]
- Cui, X.; Settleman, J.; Vasudevaraja, V.; Serrano, J.; Tong, J.; Peng, Y.; DeLorenzo, M.; Shen, G.; Frenster, J.; Morales, R.-T.T.; et al. Dissecting the immunosuppressive tumor microenvironments in Glioblastoma-on-a-Chip for optimized PD-1 immunotherapy. eLife 2020, 9, 52253. [Google Scholar] [CrossRef]
- Liu, X.; Raju, P. 5.42–In Vitro Cancer Model for Drug Testing. In Comprehensive Biotechnology, 2nd ed.; Academic Press: Burlington, NJ, USA, 2011; pp. 543–549. [Google Scholar] [CrossRef]
- Nashimoto, Y.; Okada, R.; Hanada, S.; Arima, Y.; Nishiyama, K.; Miura, T.; Yokokawa, R. Vascularized cancer on a chip: The effect of perfusion on growth and drug delivery of tumor spheroid. Biomaterials 2020, 229, 119547. [Google Scholar] [CrossRef] [PubMed]
- Dogan, E.; Kisim, A.; Bati-Ayaz, G.; Kubicek, G.J.; Pesen-Okvur, D.; Miri, A.K. Cancer Stem Cells in Tumor Modeling: Challenges and Future Directions. Adv. NanoBiomed Res. 2021, 2100017. [Google Scholar] [CrossRef]
- Ching, T.; Toh, Y.-C.; Hashimoto, M.; Zhang, Y.S. Bridging the academia-to-industry gap: Organ-on-a-chip platforms for safety and toxicology assessment. Trends Pharmacol. Sci. 2021, 42, 715–728. [Google Scholar] [CrossRef] [PubMed]
- Low, L.A.; Mummery, C.; Berridge, B.R.; Austin, C.P.; Tagle, D.A. Organs-on-chips: Into the next decade. Nat. Rev. Drug Discov. 2021, 20, 345–361. [Google Scholar] [CrossRef] [PubMed]





| Material | Relevant Property | Application | References |
|---|---|---|---|
| Teflon | Ease of fabrication with maximum chemical resistance | Very sensitive assays, ultra-clean tools, valves, and pumps fabrication | [51] |
| Acrylonitrile butadiene styrene | High resolution, best topography | Crafting of the master mold, study of pathogenic organisms | [52] |
| Styrene ethylene butylene styrene | Low drug absorption, optical transmittance | Human lung epithelial cells, human umbilical vein endothelial HUVECs, human alveolar epithelial cells | [54] |
| Chitosan | Biocompatible, effective control of stereochemistry | Biosensors, film organization | [55,56] |
| Silkworm | Biocompatible, pliable | Fabrication of microfluidic platforms | [57,58] |
| PDMS | Good turnaround time, multi-material printing, long-lasting and high-temperature-resistant substance | Master molding | [59] |
| Agarose | Minimal toxicity, biodegradability, tunable stability at a lower solid ratio | Chondrocytes, AML-12 murine hepatocytes, sensors, and actuators | [61,62] |
| Photocurable resin/polymer | Effective resolution with small characters | Study of cell growth | [63,64] |
| Polyurethane-methacrylate | Economical to manufacture, biocompatibility, no cytotoxicity, strong electroosmotic mobility | Increased-aspect-ratio microstructures | [65,66,67] |
| Polyhydroxyalkanoates | Biocompatibility, tunable, biodegradability | Microfilm barrier for vapor and oxygen | [68] |
| Polyethylene glycols | Cheaper than many of the material, different weight categories are available, biocompatible, cytotoxicity approximately naught | Microfluidic valves, microfluidic channels with an increased expiry time | [69] |
| Gelatin methacrylate | Photopolymerizable, porous membrane | Mechanistic vascular and valvular biology cell support matrix | [70] |
| Polylactic acid and polyglycolic acid | Mechanical biodegradation | Porous scaffold for cell culture with better adhesion | [71] |
| Synthetic hydrogels | Induration and contraction act as sensors and actuators | Self-regulating valves, micro-lens arrays, drug release, antigen adsorption flow sensors pH regulators | [72,73] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cecen, B.; Karavasili, C.; Nazir, M.; Bhusal, A.; Dogan, E.; Shahriyari, F.; Tamburaci, S.; Buyukoz, M.; Kozaci, L.D.; Miri, A.K. Multi-Organs-on-Chips for Testing Small-Molecule Drugs: Challenges and Perspectives. Pharmaceutics 2021, 13, 1657. https://doi.org/10.3390/pharmaceutics13101657
Cecen B, Karavasili C, Nazir M, Bhusal A, Dogan E, Shahriyari F, Tamburaci S, Buyukoz M, Kozaci LD, Miri AK. Multi-Organs-on-Chips for Testing Small-Molecule Drugs: Challenges and Perspectives. Pharmaceutics. 2021; 13(10):1657. https://doi.org/10.3390/pharmaceutics13101657
Chicago/Turabian StyleCecen, Berivan, Christina Karavasili, Mubashir Nazir, Anant Bhusal, Elvan Dogan, Fatemeh Shahriyari, Sedef Tamburaci, Melda Buyukoz, Leyla Didem Kozaci, and Amir K. Miri. 2021. "Multi-Organs-on-Chips for Testing Small-Molecule Drugs: Challenges and Perspectives" Pharmaceutics 13, no. 10: 1657. https://doi.org/10.3390/pharmaceutics13101657
APA StyleCecen, B., Karavasili, C., Nazir, M., Bhusal, A., Dogan, E., Shahriyari, F., Tamburaci, S., Buyukoz, M., Kozaci, L. D., & Miri, A. K. (2021). Multi-Organs-on-Chips for Testing Small-Molecule Drugs: Challenges and Perspectives. Pharmaceutics, 13(10), 1657. https://doi.org/10.3390/pharmaceutics13101657