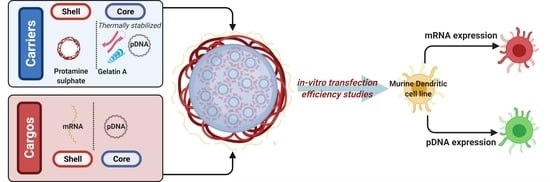
Co-Delivery of mRNA and pDNA Using Thermally Stabilized Coacervate-Based Core-Shell Nanosystems
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials for Nanocarriers and Controls
2.2. Materials for Analytics
2.3. Nucleic Acids
2.4. Cell Culture
2.5. Particle Core Assembly
2.6. Thermal Particle Core Stabilization
2.7. Shell Deposition and mRNA Loading
2.8. Dynamic Light Scattering (DLS)
2.9. Nanoparticle Tracking Analysis (NTA)
2.10. Circular Dichroism (CD)
2.11. Transmission Electron Microscopy (TEM)
2.12. Agarose Gel Electrophoresis
2.13. PicoGreen and RiboGreen Assays
2.14. In-Vitro Biological Assessment of the Nanocarrier
2.15. Confocal Laser Scanning Microscopy (CLSM)
2.16. Statistical Analysis
3. Results and Discussion
3.1. Particle Preparation
3.1.1. Core Assembly
3.1.2. Core Stabilization
3.1.3. Shell Deposition
3.2. Entrapment Efficiency and Nanocarrier NA Capacity
3.3. Cytotoxicity Assay
3.4. Transfection Efficiency of Co-Delivered mRNA (mCherry) and pDNA (pAmCyan1) in Murine Dendritic Cell Line DC2.4
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yamakawa, K.; Nakano-Narusawa, Y.; Hashimoto, N.; Yokohira, M.; Matsuda, Y. Development and clinical trials of nucleic acid medicines for pancreatic cancer treatment. Int. J. Mol. Sci. 2019, 20, 4224. [Google Scholar] [CrossRef] [Green Version]
- Sridharan, K.; Gogtay, N.J. Therapeutic nucleic acids: Current clinical status. Brit. J. Clin. Pharmacol. 2016, 82, 659–672. [Google Scholar] [CrossRef] [Green Version]
- Jackson, N.A.C.; Kester, K.E.; Casimiro, D.; Gurunathan, S.; DeRosa, F. The promise of mRNA vaccines: A biotech and industrial perspective. Vaccines 2020, 5, 11. [Google Scholar] [CrossRef]
- Redding, L.; Weiner, D.B. DNA vaccines in veterinary use. Expert Rev. Vaccines 2014, 8, 1251–1276. [Google Scholar] [CrossRef] [Green Version]
- FDA. FDA Takes Key Action in Fight Against COVID-19 By Issuing Emergency Use Authorization for First COVID-19 Vaccine. 2020. Available online: https://www.fda.gov/news-events/press-announcements/fda-takes-key-action-fight-against-covid-19-issuing-emergency-use-authorization-first-covid-19 (accessed on 30 September 2021).
- FDA. Moderna COVID-19 Vaccine. 2021. Available online: https://www.fda.gov/emergency-preparedness-and-response/coronavirus-disease-2019-covid-19/moderna-covid-19-vaccine (accessed on 30 September 2021).
- Chung, Y.H.; Beiss, V.; Fiering, S.N.; Steinmetz, N.F. COVID-19 vaccine frontrunners and their nanotechnology design. ACS Nano 2020, 14, 12522–12537. [Google Scholar] [CrossRef]
- Gupta, A.; Andresen, J.L.; Manan, R.S.; Langer, R. Nucleic acid delivery for therapeutic applications. Adv. Drug Deliv. Rev. 2021, 2021, 113834. [Google Scholar] [CrossRef]
- Khurana, B.; Goyal, A.K.; Budhiraja, A.; Aora, D.; Vyas, S.P. Lipoplexes versus nanoparticles: PDNA/SiRNA delivery. Drug Deliv. 2013, 20, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Veilleux, D.; Lavertu, M.; Biniecki, K.; Buschmann, M.D. Lyophilized chitosan nanoparticles for PDNA 766 and SiRNA delivery: Physico-chemical properties, transfection efficiency, and cytotoxicity. Mol. Ther. 2014, 22, S134. [Google Scholar] [CrossRef]
- Moradian, H.; Lendlein, A.; Gossen, M. Strategies for simultaneous and successive delivery of RNA. J. Mol. Med. 2020, 98, 1767–1779. [Google Scholar] [CrossRef]
- Ball, R.L.; Hajj, K.A.; Vizelman, J.; Bajaj, P.; Whitehead, K.A. Lipid nanoparticle formulations for enhanced co-delivery of SiRNA and mRNA. Nano Lett. 2018, 18, 3814–3822. [Google Scholar] [CrossRef] [PubMed]
- Lundstrom, K. Latest development on RNA-based drugs and vaccines. Future Sci. OA 2018, 4, FSO300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karikó, K.; Muramatsu, H.; Welsh, F.A.; Ludwig, J.; Kato, H.; Akira, S.; Weissman, D. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. 2008, 16, 1833–1840. [Google Scholar] [CrossRef]
- Baiersdörfer, M.; Boros, G.; Muramatsu, H.; Mahiny, A.; Vlatkovic, I.; Sahin, U.; Karikó, K. A facile method for the removal of dsRNA Contaminant from in vitro-transcribed mRNA. Mol. Ther. Nucleic Acids 2019, 15, 26–35. [Google Scholar] [CrossRef] [Green Version]
- Beuckelaer, A.D.; Pollard, C.; Lint, S.V.; Roose, K.; Hoecke, L.V.; Naessens, T.; Udhayakumar, V.K.; Smet, M.; Sanders, N.; Lienenklaus, S.; et al. Type I interferons interfere with the capacity of mRNA lipoplex vaccines to elicit cytolytic T cell responses. Mol. Ther. 2016, 24, 2012–2020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Devoldere, J.; Dewitte, H.; Smedt, S.C.D.; Remaut, K. Evading innate immunity in nonviral mRNA delivery: Don’t shoot the messenger. Drug Discov. Today 2016, 21, 11–25. [Google Scholar] [CrossRef] [Green Version]
- Karikó, K.; Buckstein, M.; Ni, H.; Weissman, D. Suppression of RNA recognition by toll-like receptors: The impact of nucleoside modification and the evolutionary origin of RNA. Immunity 2005, 23, 165–175. [Google Scholar] [CrossRef] [Green Version]
- Kormann, M.S.D.; Hasenpusch, G.; Aneja, M.K.; Nica, G.; Flemmer, A.W.; Herber-Jonat, S.; Huppmann, M.; Mays, L.E.; Illenyi, M.; Schams, A.; et al. Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat. Biotechnol. 2011, 29, 154–157. [Google Scholar] [CrossRef]
- Verbeke, R.; Lentacker, I.; Wayteck, L.; Breckpot, K.; Bockstal, M.V.; Descamps, B.; Vanhove, C.; Smedt, S.C.D.; Dewitte, H. Co-delivery of nucleoside-modified mRNA and TLR agonists for cancer immunotherapy: Restoring the immunogenicity of immunosilent mRNA. J. Control Release 2017, 266, 287–300. [Google Scholar] [CrossRef] [PubMed]
- Linares-Fernández, S.; Lacroix, C.; Exposito, J.-Y.; Verrier, B. Tailoring mRNA vaccine to balance innate/adaptive immune response. Trends Mol. Med. 2019, 26, 311–323. [Google Scholar] [CrossRef]
- Minnaert, A.-K.; Vanluchene, H.; Verbeke, R.; Lentacker, I.; Smedt, S.C.D.; Raemdonck, K.; Sanders, N.; Remaut, K. Strategies for controlling the innate immune activity of conventional and self-amplifying mRNA therapeutics: Getting the message across. Adv. Drug Deliv. Rev. 2021, 176, 113900. [Google Scholar] [CrossRef]
- Hotz, C.; Wagenaar, T.R.; Gieseke, F.; Bangari, D.S.; Callahan, M.; Cao, H.; Diekmann, J.; Diken, M.; Grunwitz, C.; Hebert, A.; et al. Local delivery of mRNA-encoding cytokines promotes antitumor immunity and tumor eradication across multiple preclinical tumor models. Sci. Transl. Med. 2021, 13, eabc7804. [Google Scholar] [CrossRef] [PubMed]
- Dewitte, H.; Verbeke, R.; Breckpot, K.; Smedt, S.C.D.; Lentacker, I. Nanoparticle design to induce tumor immunity and challenge the suppressive tumor microenvironment. Nano Today 2014, 9, 743–758. [Google Scholar] [CrossRef] [Green Version]
- Kreutz, M.; Giquel, B.; Hu, Q.; Abuknesha, R.; Uematsu, S.; Akira, S.; Nestle, F.O.; Diebold, S.S. Antibody-antigen-adjuvant conjugates enable co-delivery of antigen and adjuvant to dendritic cells in cis but only have partial targeting specificity. PLoS ONE 2012, 7, e40208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, B.; Zhao, J.; Li, H.; He, K.-L.; Chen, Y.; Chen, S.-H.; Mayer, L.; Unkeless, J.C.; Xiong, H. Toll-like receptors on tumor cells facilitate evasion of immune surveillance. Cancer Res. 2005, 65, 5009–5014. [Google Scholar] [CrossRef] [Green Version]
- Chiodoni, C.; Iezzi, M.; Guiducci, C.; Sangaletti, S.; Alessandrini, I.; Ratti, C.; Tiboni, F.; Musiani, P.; Granger, D.N.; Colombo, M.P. Triggering CD40 on endothelial cells contributes to tumor growth. J. Exp. Med. 2006, 203, 2441–2450. [Google Scholar] [CrossRef] [Green Version]
- Lint, S.V.; Renmans, D.; Broos, K.; Goethals, L.; Maenhout, S.; Benteyn, D.; Goyvaerts, C.; Four, S.D.; der Jeught, K.V.; Bialkowski, L.; et al. Intratumoral delivery of TriMix mRNA results in T-cell activation by cross-presenting dendritic cells. Cancer Immunol. Res. 2016, 4, 146–156. [Google Scholar] [CrossRef] [Green Version]
- Keersmaecker, B.D.; Claerhout, S.; Carrasco, J.; Bar, I.; Corthals, J.; Wilgenhof, S.; Neyns, B.; Thielemans, K. TriMix and tumor antigen mRNA electroporated dendritic cell vaccination plus ipilimumab: Link between T-cell activation and clinical responses in advanced melanoma. J. Immunother. Cancer 2020, 8, e000329. [Google Scholar] [CrossRef] [Green Version]
- Van der Aa, M.A.E.M.; Mastrobattista, E.; Oosting, R.S.; Hennink, W.E.; Koning, G.A.; Crommelin, D.J.A. The nuclear pore complex: The gateway to successful nonviral gene delivery. Pharm. Res. 2006, 23, 447–459. [Google Scholar] [CrossRef]
- Ross, R.; Sudowe, S.; Beisner, J.; Ross, X.-L.; Ludwig-Portugall, I.; Steitz, J.; Tüting, T.; Knop, J.; Reske-Kunz, A.B. Transcriptional targeting of dendritic cells for gene therapy using the promoter of the cytoskeletal protein fascin. Gene Ther. 2003, 10, 1035–1040. [Google Scholar] [CrossRef] [Green Version]
- Moulin, V.; Morgan, M.E.; Eleveld-Trancikova, D.; Haanen, J.B.A.G.; Wielders, E.; Looman, M.W.G.; Janssen, R.A.J.; Figdor, C.G.; Jansen, B.J.H.; Adema, G.J. Targeting dendritic cells with antigen via dendritic cell-associated promoters. Cancer Gene Ther. 2012, 19, 303–311. [Google Scholar] [CrossRef] [Green Version]
- Jones, D.L.; Brewster, R.C.; Phillips, R. Promoter architecture dictates cell-to-cell variability in gene expression. Science 2014, 346, 1533–1536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andreev, D.E.; Terenin, I.M.; Dmitriev, S.E.; Shatsky, I.N. Pros and cons of PDNA and mRNA transfection to study mRNA translation in mammalian cells. Gene 2016, 578, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Bauerschmitz, G.J.; Nettelbeck, D.M.; Kanerva, A.; Baker, A.H.; Hemminki, A.; Reynolds, P.N.; Curiel, D.T. The Flt-1 promoter for transcriptional targeting of teratocarcinoma. Cancer Res. 2002, 62, 1271–1274. [Google Scholar]
- Morán, M.C.; Forniés, I.; Ruano, G.; Busquets, M.A.; Vinardell, M.P. Efficient encapsulation and release of RNA molecules from gelatin-based nanoparticles. Colloids Surf. Physicochem. Eng. Asp. 2017, 516, 226–237. [Google Scholar] [CrossRef]
- Gorgieva, S.; Kokol, V. Collagen- vs. gelatine-based biomaterials and their biocompatibility: Review and perspectives. In Biomaterials Applications for Nanomedicine; IntechOpen: London, UK, 2011. [Google Scholar]
- Noguchi, A.; Hirashima, N.; Nakanishi, M. Cationic cholesterol promotes gene transfection using the nuclear localization signal in protamine. Pharm. Res. 2002, 19, 933–938. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Chen, S.; Jiang, Y.; Jiang, J.; Zhang, Z.; Sun, X. Dendritic cell targeted liposomes–protamine–DNA complexes mediated by synthetic mannosylated cholestrol as a potential carrier for DNA vaccine. Nanotechnology 2013, 24, 295101. [Google Scholar] [CrossRef]
- Nafee, N.; Schneider, M.; Lehr, C.-M. Multifunctional pharmaceutical nanocarriers. Fundam. Biomed. Technol. 2008, 337–362. [Google Scholar] [CrossRef]
- Zagato, E.; Vermeulen, L.; Dewitte, H.; Imschoot, G.V.; Vandenbroucke, R.E.; Demeester, J.; Smedt, S.C.D.; Neyts, K.; Remaut, K.; Braeckmans, K. Quantifying the average number of nucleic acid therapeutics per nanocarrier by single particle tracking microscopy. Mol. Pharm. 2018, 15, 1142–1149. [Google Scholar] [CrossRef]
- Schoenmaker, L.; Witzigmann, D.; Kulkarni, J.A.; Verbeke, R.; Kersten, G.; Jiskoot, W.; Crommelin, D. mRNA-lipid nanoparticle COVID-19 vaccines: Structure and stability. Int. J. Pharm. 2021, 601, 120586. [Google Scholar] [CrossRef]
- Arfin, N.; Aswal, V.K.; Bohidar, H.B. Overcharging, thermal, viscoelastic and hydration properties of DNA–gelatin complex coacervates: Pharmaceutical and food industries. RSC Adv. 2014, 4, 11705–11713. [Google Scholar] [CrossRef]
- Kumar, A.; Dixit, C.K. Methods for characterization of nanoparticles. Adv. Nanomed. Deliv. Ther. Nucleic Acids 2017, 43–58. [Google Scholar] [CrossRef]
- Burgess, D.J.; Carless, J.E. Manufacture of gelatin/gelatin coacervate microcapsules. Int. J. Pharm. 1985, 27, 61–70. [Google Scholar] [CrossRef]
- Geggier, S.; Kotlyar, A.; Vologodskii, A. Temperature dependence of DNA persistence length. Biophys. J. 2011, 100, 76a. [Google Scholar] [CrossRef] [Green Version]
- Rawat, K.; Pathak, J.; Bohidar, H.B. Effect of persistence length on binding of DNA to polyions and overcharging of their intermolecular complexes in aqueous and in 1-methyl-3-octyl imidazolium chloride ionic liquid solutions. Phys. Chem. Chem. Phys. 2013, 15, 12262–12273. [Google Scholar] [CrossRef]
- Mao, W.; Gao, Q.; Liu, Y.; Fan, Y.; Hu, L.; Xu, H. Temperature dependence of DNA condensation at high ionic concentration. Mod. Phys. Lett. B 2016, 30, 1650298. [Google Scholar] [CrossRef]
- Gornall, J.L.; Terentjev, E.M. Helix–coil transition of gelatin: Helical morphology and stability. Soft Matter 2008, 4, 544–549. [Google Scholar] [CrossRef] [PubMed]
- Gopal, R.; Park, J.S.; Seo, C.H.; Park, Y. Applications of circular dichroism for structural analysis of gelatin and antimicrobial peptides. Int. J. Mol. Sci. 2012, 13, 3229–3244. [Google Scholar] [CrossRef] [PubMed]
- Mohiti-Asli, M.; Loboa, E.G. Nanofibrous smart bandages for wound care. Wound Health Biomater. 2016, 2, 483–499. [Google Scholar] [CrossRef]
- Hellmund, M.; Achazi, K.; Neumann, F.; Thota, B.N.S.; Ma, N.; Haag, R. Systematic adjustment of charge densities and size of polyglycerol amines reduces cytotoxic effects and enhances cellular uptake. Biomater. Sci. 2015, 3, 1459–1465. [Google Scholar] [CrossRef]
- Kommareddy, S.; Amiji, M. Poly(ethylene glycol)–modified thiolated gelatin nanoparticles for glutathione-responsive intracellular DNA delivery. Nanomed. Nanotechnol. Biol. Med. 2007, 3, 32–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zwiorek, K.; Bourquin, C.; Battiany, J.; Winter, G.; Endres, S.; Hartmann, G.; Coester, C. Delivery by cationic gelatin nanoparticles strongly increases the immunostimulatory effects of CpG oligonucleotides. Pharm. Res. 2008, 25, 551–562. [Google Scholar] [CrossRef]
- Vaz, B.; Popovic, M.; Ramadan, K. DNA–protein crosslink proteolysis repair. Trends Biochem. Sci. 2017, 42, 483–495. [Google Scholar] [CrossRef]
- Klages-Mundt, N.L.; Li, L. Formation and repair of DNA-protein crosslink damage. Sci. China Life Sci. 2017, 60, 1065–1076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tretyakova, N.Y.; Groehler, A.; Ji, S. DNA–protein cross-links: Formation, structural identities, and biological outcomes. Acc. Chem. Res. 2015, 48, 1631–1644. [Google Scholar] [CrossRef] [Green Version]
- Rawat, K.; Aswal, V.K.; Bohidar, H.B. DNA–gelatin complex coacervation, UCST and first-order phase transition of coacervate to anisotropic ion gel in 1-methyl-3-octylimidazolium chloride ionic liquid solutions. J. Phys. Chem. B 2012, 116, 14805–14816. [Google Scholar] [CrossRef]
- Kamla, R.; Bohidar, H. Coacervation in biopolymers. J. Phys. Chem. Biophys. 2014, 4, 1. [Google Scholar] [CrossRef] [Green Version]
- Yang, G.; Xiao, Z.; Long, H.; Ma, K.; Zhang, J.; Ren, X.; Zhang, J. Assessment of the characteristics and biocompatibility of gelatin sponge scaffolds prepared by various crosslinking methods. Sci. Rep. 2018, 8, 1616. [Google Scholar] [CrossRef] [PubMed]
- Ulubayram, K.; Aksu, E.; Gurhan, S.I.D.; Serbetci, K.; Hasirci, N. Cytotoxicity evaluation of gelatin sponges prepared with different cross-linking agents. J. Biomater. Sci. 2002, 13, 1203–1219. [Google Scholar] [CrossRef] [Green Version]
- Sebastian, M.; Schröder, A.; Scheel, B.; Hong, H.S.; Muth, A.; von Boehmer, L.; Zippelius, A.; Mayer, F.; Reck, M.; Atanackovic, D.; et al. A phase I/IIa study of the mRNA-based cancer immunotherapy CV9201 in patients with stage IIIB/IV non-small cell lung cancer. Cancer Immunol. Immunother. 2019, 68, 799–812. [Google Scholar] [CrossRef]
- Kübler, H.; Scheel, B.; Gnad-Vogt, U.; Miller, K.; Schultze-Seemann, W.; vom Dorp, F.; Parmiani, G.; Hampel, C.; Wedel, S.; Trojan, L.; et al. Self-adjuvanted mRNA vaccination in advanced prostate cancer patients: A first-in-man phase I/IIa study. J. Immunother. Cancer 2015, 3, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kallen, K.-J.; Heidenreich, R.; Schnee, M.; Petsch, B.; Schlake, T.; Thess, A.; Baumhof, P.; Scheel, B.; Koch, S.D.; Fotin-Mleczek, M. A novel, disruptive vaccination technology. Hum. Vacc. Immunother. 2013, 9, 2263–2276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alberer, M.; Gnad-Vogt, U.; Hong, H.S.; Mehr, K.T.; Backert, L.; Finak, G.; Gottardo, R.; Bica, M.A.; Garofano, A.; Koch, S.D.; et al. Safety and immunogenicity of a mRNA rabies vaccine in healthy adults: An open-label, non-randomised, prospective, first-in-human phase 1 clinical trial. Lancet 2017, 390, 1511–1520. [Google Scholar] [CrossRef]
- Papachristofilou, A.; Hipp, M.M.; Klinkhardt, U.; Früh, M.; Sebastian, M.; Weiss, C.; Pless, M.; Cathomas, R.; Hilbe, W.; Pall, G.; et al. Phase Ib evaluation of a self-adjuvanted protamine formulated mRNA-based active cancer immunotherapy, BI1361849 (CV9202), combined with local radiation treatment in patients with stage IV non-small cell lung cancer. J. Immunother. Cancer 2019, 7, 38. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, F.; Weissleder, R.; Josephson, L. Protamine as an efficient membrane-translocating peptide. Bioconjugate Chem. 2005, 16, 1240–1245. [Google Scholar] [CrossRef] [PubMed]
- Tenkumo, T.; Rotan, O.; Sokolova, V.; Epple, M. Protamine increases transfection efficiency and cell viability after transfection with calcium phosphate nanoparticles. Nano Biomed. 2014, 5, 64–74. [Google Scholar] [CrossRef]
- Masuda, T.; Akita, H.; Harashima, H. Evaluation of nuclear transfer and transcription of plasmid DNA condensed with protamine by microinjection: The use of a nuclear transfer score. FEBS Lett. 2005, 579, 2143–2148. [Google Scholar] [CrossRef] [Green Version]
- Sorgi, F.; Bhattacharya, S.; Huang, L. Protamine sulfate enhances lipid-mediated gene transfer. Gene Ther. 1997, 4, 961–968. [Google Scholar] [CrossRef]







| Sample | Gelatin Concentration [mg·mL−1] | DNA Concentration [µg·mL−1] | Gelatin to DNA Ratio [w/w] | Protamine Sulphate Concentration [mg·mL−1] | Protamine Sulphate to Gelatin Ratio [w/w] |
|---|---|---|---|---|---|
| CoAc100 | 10 | 100 | 100:1 | – | – |
| CoAc70 | 7 | 100 | 70:1 | – | – |
| CoAc50 | 5 | 100 | 50:1 | – | – |
| CoAc30 1 | 3 | 100 | 30:1 | – | – |
| CoAc20 | 2 | 100 | 20:1 | – | – |
| CoAc1 | 0.1 | 100 | 1:1 | – | – |
| TS-CoAc 2 | 3 | 100 | 30:1 | – | – |
| P-CoAc 3 | 3 | 100 | 30:1 | 0.3 | 1:5 |
| P-TS-CoAc 2,3 | 3 | 100 | 30:1 | 0.3 | 1:5 |
| pAmCyan | mCherry | |||||
|---|---|---|---|---|---|---|
| Sample | EE [%] | Molecules/Dose | Molecules/NP | EE [%] | Molecules/Dose | Molecules/NP |
| CoAc | 100.10 ± 0.28% | 1.076 × 1012 | 5318 | No colloidally stable coated P-CoAc for surface-loading | ||
| (P-)TS-CoAc | 100.12 ± 0.39% | 1.076 × 1012 | 5318 | 97.81 ± 1.06 | 1.884 × 1012 | 9312 |
| jetPrime a | 100.01 ± 9 × 10−5% | 1.076 × 1012 | - | - | - | - |
| JetMessenger b | - | - | - | 100.66 ± 20.94% | 1.884 × 1012 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nasr, S.S.; Lee, S.; Thiyagarajan, D.; Boese, A.; Loretz, B.; Lehr, C.-M. Co-Delivery of mRNA and pDNA Using Thermally Stabilized Coacervate-Based Core-Shell Nanosystems. Pharmaceutics 2021, 13, 1924. https://doi.org/10.3390/pharmaceutics13111924
Nasr SS, Lee S, Thiyagarajan D, Boese A, Loretz B, Lehr C-M. Co-Delivery of mRNA and pDNA Using Thermally Stabilized Coacervate-Based Core-Shell Nanosystems. Pharmaceutics. 2021; 13(11):1924. https://doi.org/10.3390/pharmaceutics13111924
Chicago/Turabian StyleNasr, Sarah S., Sangeun Lee, Durairaj Thiyagarajan, Annette Boese, Brigitta Loretz, and Claus-Michael Lehr. 2021. "Co-Delivery of mRNA and pDNA Using Thermally Stabilized Coacervate-Based Core-Shell Nanosystems" Pharmaceutics 13, no. 11: 1924. https://doi.org/10.3390/pharmaceutics13111924
APA StyleNasr, S. S., Lee, S., Thiyagarajan, D., Boese, A., Loretz, B., & Lehr, C. -M. (2021). Co-Delivery of mRNA and pDNA Using Thermally Stabilized Coacervate-Based Core-Shell Nanosystems. Pharmaceutics, 13(11), 1924. https://doi.org/10.3390/pharmaceutics13111924