Efficacy of Ursolic Acid-Enriched Water-Soluble and Not Cytotoxic Nanoparticles against Enterococci
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals Substances and Instruments Used in This Study
2.2. Microbiologic Investigations
2.2.1. Description of the Microorganisms Considered in This Study
2.2.2. Definition of the Minimal Inhibitory Concentrations (MICs)
2.2.3. Time-Killing Experiments
2.3. Evaluation of UA, G4K and UA-G4K NPs Cytotoxicity on Eukaryotic Normal Cells
2.3.1. The Cell Culture Used in This Study
2.3.2. Assessment of Viability of HaCaT Cells Exposed to G4K, UA, and UA-G4K NPs
2.4. Statistical Analyses
3. Results
3.1. Synthesis and Characterization of UA-G4K NPs
3.2. Antibacterial Properties of UA and UA-G4K NPs
3.2.1. Determination of MICs of UA-G4K and UA
3.2.2. Relevance of Our Results
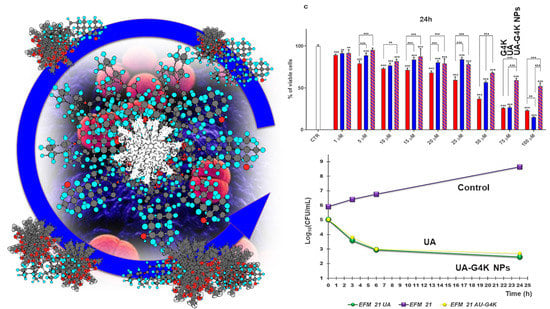
3.2.3. Curves from Time-Killing Experiments
3.3. Cytotoxicity Effects of G4K, UA and UA-G4K NPs on HaCaT Human Keratinocytes Cells
Dose- and Time-Dependent Cytotoxicity Experiments
4. Conclusions and Future Perspectives for UA-G4K NPs
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization (WHO). No Time to Wait: Securing the Future from Drug-Resistant Infections. Report to the Secretary-General of the United Nations. Interagency Coordination Group on Antimicrobial Resistance; WHO: Geneva, Switzerland, 2019; Available online: https://www.who.int/antimicrobial-resistance/interagency-coordination-group/IACG_final_report_EN.pdf?ua=1 (accessed on 29 October 2021).
- Woźniak, Ł.; Skąpska, S.; Marszałek, K. Ursolic Acid—A Pentacyclic Triterpenoid with a Wide Spectrum of Pharmacological Activities. Molecules 2015, 20, 20614–20641. [Google Scholar] [CrossRef] [Green Version]
- Gazzarata, R.; Monteverde, M.E.; Ruggiero, C.; Maggi, N.; Palmieri, D.; Parruti, G.; Giacomini, M. Healthcare Associated Infections: An Interoperable Infrastructure for Multidrug Resistant Organism Surveillance. IJERPH 2020, 17, 465. [Google Scholar] [CrossRef] [Green Version]
- Burnham, J.P.; Olsen, M.A.; Kollef, M.H. Re-Estimating Annual Deaths Due to Multidrug-Resistant Organism Infections. Infect. Control. Hosp. Epidemiol. 2019, 40, 112–113. [Google Scholar] [CrossRef] [Green Version]
- de Kraker, M.E.A.; Stewardson, A.J.; Harbarth, S. Will 10 Million People Die a Year Due to Antimicrobial Resistance by 2050? PLoS Med. 2016, 13, e1002184. [Google Scholar] [CrossRef] [Green Version]
- Limmathurotsakul, D.; Dunachie, S.; Fukuda, K.; Feasey, N.A.; Okeke, I.N.; Holmes, A.H.; Moore, C.E.; Dolecek, C.; van Doorn, H.R.; Shetty, N.; et al. Improving the Estimation of the Global Burden of Antimicrobial Resistant Infections. Lancet Infect. Dis. 2019, 19, e392–e398. [Google Scholar] [CrossRef]
- Denkinger, C.M.; Grant, A.D.; Denkinger, M.; Gautam, S.; D’Agata, E.M.C. Increased Multi-Drug Resistance among the Elderly on Admission to the Hospital—A 12-Year Surveillance Study. Arch. Gerontol. Geriatr. 2013, 56, 227–230. [Google Scholar] [CrossRef] [PubMed]
- Burke, J.P. Infection Control—A Problem for Patient Safety. N. Engl. J. Med. 2003, 348, 651–656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abreu, A.C.; Borges, A.; Malheiro, J.; Simões, M. Resurgence of the interest in plants as sources of medicines and resistance-modifying agents. In Microbial Pathogens and Strategies for Combating Them: Science, Technology and Education, 1st ed.; Méndez-Vilas, A., Ed.; Formatex Research Center, S.L.: Badajoz, Spain, 2013; Volume 2, pp. 1287–1297. [Google Scholar]
- Koulenti, D.; Xu, E.; Mok, I.Y.S.; Song, A.; Karageorgopoulos, D.E.; Armaganidis, A.; Lipman, J.; Tsiodras, S. Novel Antibiotics for Multidrug-Resistant Gram-Positive Microorganisms. Microorganisms 2019, 7, 270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Q.; He, M.; Zhang, M.; Zeng, S.; Chen, L.; Zhou, L.; Xu, H. Ursolic acid: A systematic review of its pharmacology, toxicity and rethink on its pharmacokinetics based on PK-PD model. Fitoterapia 2020, 147, 104735. [Google Scholar] [CrossRef]
- Fan, J.-P.; Liao, D.-D.; Zhang, X.-H. Ultrasonic Assisted Extraction of Ursolic Acid from Apple Pomace: A Novel and Facile Technique. Sep. Sci. Technol. 2016, 51, 1344–1350. [Google Scholar] [CrossRef]
- Fan, J.-P.; Cao, Y.-H.; Zhang, X.-H.; Jiang, D.-Q.; Yu, J.-X. Determination and Modeling of the Solubilities of Oleanolic Acid and Ursolic Acid in Ethanol + Sodium Hydroxide + Water Mixed Solvents from T = 283.2 to 323.2 K. J. Chem. Eng. Data 2017, 62, 3991–3997. [Google Scholar] [CrossRef]
- Fan, J.-P.; Kong, T.; Zhang, X.-H.; Zhang, L.; Tong, S.; Tian, Z.-Y.; Zhu, J.-H. Solubilities of Oleanolic Acid and Ursolic Acid in (Ethanol + water) Mixed Solvents from T = (292.2 to 328.2) K. J. Chem. Thermodyn. 2012, 47, 372–375. [Google Scholar] [CrossRef]
- Fan, J.-P.; Lai, X.-H.; Tian, X.; Zhang, X.-H.; Cao, Y.-H.; Chen, H.-P. Solubilities of Oleanolic Acid and Ursolic Acid in Different Organic Solvents and 2-Propanol + Water Binary Solvent Mixtures at Different Temperatures: Experimental Measurement and Modeling. J. Chem. Eng. Data 2021, 66, 684–691. [Google Scholar] [CrossRef]
- Kashyap, D.; Tuli, H.S.; Sharma, A.K. Ursolic Acid (UA): A Metabolite with Promising Therapeutic Potential. Life Sci. 2016, 146, 201–213. [Google Scholar] [CrossRef]
- Seo, D.Y.; Lee, S.R.; Heo, J.-W.; No, M.-H.; Rhee, B.D.; Ko, K.S.; Kwak, H.-B.; Han, J. Ursolic Acid in Health and Disease. Korean J. Physiol. Pharmacol. 2018, 22, 235. [Google Scholar] [CrossRef] [Green Version]
- López-Hortas, L.; Pérez-Larrán, P.; González-Muñoz, M.J.; Falqué, E.; Domínguez, H. Recent Developments on the Extraction and Application of Ursolic Acid. A Review. Food Res. Int. 2018, 103, 130–149. [Google Scholar] [CrossRef]
- Jäger, S.; Trojan, H.; Kopp, T.; Laszczyk, M.; Scheffler, A. Pentacyclic Triterpene Distribution in Various Plants—Rich Sources for a New Group of Multi-Potent Plant Extracts. Molecules 2009, 14, 2016–2031. [Google Scholar] [CrossRef] [Green Version]
- Baricevic, D.; Sosa, S.; Della Loggia, R.; Tubaro, A.; Simonovska, B.; Krasna, A.; Zupancic, A. Topical Anti-Inflammatory Activity of Salvia Officinalis L. Leaves: The Relevance of Ursolic Acid. J. Ethnopharmacol. 2001, 75, 125–132. [Google Scholar] [CrossRef]
- Bisio, A.; Fraternale, D.; Schito, A.M.; Parricchi, A.; Dal Piaz, F.; Ricci, D.; Giacomini, M.; Ruffoni, B.; De Tommasi, N. Establishment and Analysis of in Vitro Biomass from Salvia Corrugata Vahl. and Evaluation of Antimicrobial Activity. Phytochemistry 2016, 122, 276–285. [Google Scholar] [CrossRef] [PubMed]
- Bisio, A.; De Mieri, M.; Milella, L.; Schito, A.M.; Parricchi, A.; Russo, D.; Alfei, S.; Lapillo, M.; Tuccinardi, T.; Hamburger, M.; et al. Antibacterial and Hypoglycemic Diterpenoids from Salvia Chamaedryoides. J. Nat. Prod. 2017, 80, 503–514. [Google Scholar] [CrossRef]
- Bisio, A.; Romussi, G.; Russo, E.; Cafaggi, S.; Schito, A.M.; Repetto, B.; De Tommasi, N. Antimicrobial Activity of the Ornamental Species Salvia Corrugata, a Potential New Crop for Extractive Purposes. J. Agric. Food Chem. 2008, 56, 10468–10472. [Google Scholar] [CrossRef]
- Bisio, A.; Schito, A.M.; Ebrahimi, S.N.; Hamburger, M.; Mele, G.; Piatti, G.; Romussi, G.; Dal Piaz, F.; De Tommasi, N. Antibacterial Compounds from Salvia Adenophora Fernald (Lamiaceae). Phytochemistry 2015, 110, 120–132. [Google Scholar] [CrossRef]
- Fontanay, S.; Grare, M.; Mayer, J.; Finance, C.; Duval, R.E. Ursolic, oleanolic and betulinic acids: Antibacterial spectra and selectivity indexes. J. Ethnopharmacol. 2008, 120, 272–276. [Google Scholar] [CrossRef]
- Bisio, A.; Schito, A.M.; Parricchi, A.; Mele, G.; Romussi, G.; Malafronte, N.; Oliva, P.; Tommasi, N.D. Antibacterial Activity of Constituents from Salvia Buchananii Hedge (Lamiaceae). Phytochem. Lett. 2015, 14, 170–177. [Google Scholar] [CrossRef]
- Jabeen, M.; Ahmad, S.; Shahid, K.; Sadiq, A.; Rashid, U. Ursolic Acid Hydrazide Based Organometallic Complexes: Synthesis, Characterization, Antibacterial, Antioxidant, and Docking Studies. Front. Chem. 2018, 6, 55. [Google Scholar] [CrossRef]
- Kim, S.-G.; Kim, M.-J.; Jin, D.-C.; Park, S.-N.; Cho, E.-G.; Freire, M.O.; Jang, S.-J.; Park, Y.-J.; Kook, J.-K. Antimicrobial Effect of Ursolic Acid and Oleanolic Acid against Methicillin-Resistant Staphylococcus aureus. Korean J. Microbiol. 2012, 48, 212–215. [Google Scholar] [CrossRef] [Green Version]
- Jiménez-Arellanes, A.; Luna-Herrera, J.; Cornejo-Garrido, J.; López-García, S.; Castro-Mussot, M.E.; Meckes-Fischer, M.; Mata-Espinosa, D.; Marquina, B.; Torres, J.; Hernández-Pando, R. Ursolic and Oleanolic Acids as Antimicrobial and Immunomodulatory Compounds for Tuberculosis Treatment. BMC Complement. Altern. Med. 2013, 13, 258. [Google Scholar] [CrossRef] [Green Version]
- Jesus, J.A.; Lago, J.H.G.; Laurenti, M.D.; Yamamoto, E.S.; Passero, L.F.D. Antimicrobial Activity of Oleanolic and Ursolic Acids: An Update. Evid. -Based Complementary Altern. Med. 2015, 2015, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.-M.; Chen, H.-T.; Wu, Z.-Y.; Jhan, Y.-L.; Shyu, C.-L.; Chou, C.-H. Antibacterial and Synergistic Activity of Pentacyclic Triterpenoids Isolated from Alstonia Scholaris. Molecules 2016, 21, 139. [Google Scholar] [CrossRef] [Green Version]
- Chung, P.; Navaratnam, P.; Chung, L. Synergistic Antimicrobial Activity between Pentacyclic Triterpenoids and Antibiotics against Staphylococcus Aureus Strains. Ann. Clin. Microbiol. Antimicrob. 2011, 10, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, T.; Li, Z.; Kang, O.-H.; Mun, S.-H.; Seo, Y.-S.; Kong, R.; Shin, D.-W.; Liu, X.-Q.; Kwon, D.-Y. Antimicrobial Activity and Synergism of Ursolic Acid 3-O-α-L-Arabinopyranoside with Oxacillin against Methicillin-Resistant Staphylococcus Aureus. Int. J. Mol. Med. 2017, 40, 1285–1293. [Google Scholar] [CrossRef] [Green Version]
- Jyothi, J.S.; Putty, K.; Reddy, Y.N.; Dhanalakshmi, K.; Umair, M.A.H. Antagonistic Effect of Ursolic Acid on Staphylococcal Biofilms. Vet. World 2018, 11, 1440–1444. [Google Scholar] [CrossRef] [Green Version]
- Qin, N.; Tan, X.; Jiao, Y.; Liu, L.; Zhao, W.; Yang, S.; Jia, A. RNA-Seq-based transcriptome analysis of methicillin-resistant Staphylococcus aureus biofilm inhibition by ursolic acid and resveratrol. Sci. Rep. 2014, 4, 5467. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.M.; Jhan, Y.L.; Tsai, S.J.; Chou, C.H. The Pleiotropic Antibacterial Mechanisms of Ursolic Acid against Methicillin-Resistant Staphylococcus aureus (MRSA). Molecules 2016, 21, 884. [Google Scholar] [CrossRef] [Green Version]
- Maggi, N.; Schito, A.M.; Iobbi, V.; Bisio, A.; Ruggiero, C.; Giacomini, M. Molecular docking of ursolic acid and Staphylococcus aureus ATPase for antibacterial therapy. In Proceedings of the 2020 IEEE 20th Mediterranean Electrotechnical Conference (MELECON), Palermo, Italy, 16–18 July 2020; IEEE: Piscataway, NJ, USA, 2020; pp. 372–375. [Google Scholar]
- Zhang, H.; Li, X.; Ding, J.; Xu, H.; Dai, X.; Hou, Z.; Zhang, K.; Sun, K.; Sun, W. Delivery of ursolic acid (UA) in polymeric nanoparticles effectively promotes the apoptosis of gastric cancer cells through enhanced inhibition of cyclooxygenase 2 (COX-2). Int. J. Pharm. 2013, 441, 261–268. [Google Scholar] [CrossRef]
- Song, S.; Gao, K.; Niu, R.; Yi, W.; Zhang, J.; Gao, C.; Yang, B.; Liao, X. Binding behavior, water solubility and in vitro cytotoxicity of inclusion complexes between ursolic acid and amino-appended β-cyclodextrins. J. Mol. Liq. 2019, 296, 111993. [Google Scholar] [CrossRef]
- Meng, Y.Q.; Xu, C.D.; Yu, T.T.; Li, W.; Li, Q.W.; Li, X.X. Synthesis, and antitumor activity evaluation of ursolic acid derivatives. J. Asian Nat. Prod. Res. 2020, 22, 359–369. [Google Scholar] [CrossRef]
- Zhang, N.; Liu, S.; Shi, S.; Chen, Y.; Xu, F.; Wei, X.; Xu, Y. Solubilization and delivery of Ursolic-acid for modulating tumor microenvironment and regulatory T cell activities in cancer immunotherapy. J. Control. Release 2020, 320, 168–178. [Google Scholar] [CrossRef]
- Soica, C.; Oprean, C.; Borcan, F.; Danciu, C.; Trandafirescu, C.; Coricovac, D.; Crainiceanu, Z.; Dehelean, C.A.; Munteanu, M. The synergistic biologic activity of oleanolic and ursolic acids in complex with hydroxypropyl-gamma-cyclodextrin. Molecules 2014, 19, 4924–4940. [Google Scholar] [CrossRef] [Green Version]
- Cerga, O.; Borcan, F.; Ambrus, R.; Popovici, I. Syntheses of new cyclodextrin complexes with oleanolic and ursolic acids. J. Agroaliment. Process Technol. 2011, 17, 405–409. [Google Scholar]
- Wicki, A.; Witzigmann, D.; Balasubramanian, V.; Huwyler, J. Nanomedicine in cancer therapy: Challenges, opportunities, and clinical applications. J. Control. Release 2015, 200, 138–157. [Google Scholar] [CrossRef]
- Reddy, L.H.; Couvreur, P. Nanotechnology for therapy and imaging of liver diseases. J. Hepatol. 2011, 55, 1461–1466. [Google Scholar] [CrossRef] [Green Version]
- Zuccari, G.; Baldassari, S.; Alfei, S.; Marengo, B.; Valenti, G.E.; Domenicotti, C.; Ailuno, G.; Villa, C.; Marchitto, L.; Caviglioli, G. D-α-Tocopherol-Based Micelles for Successful Encapsulation of Retinoic Acid. Pharmaceuticals 2021, 14, 212. [Google Scholar] [CrossRef] [PubMed]
- Alfei, S.; Zuccari, G.; Schito, A.M. Considerable Improvement of Ursolic Acid Water Solubility by its Encapsulation in Dendrimer Nanoparticles: Design, Synthesis and Physicochemical Characterization. Nanomaterials 2021, 11. [Google Scholar] [CrossRef]
- Kristich, C.J.; Rice, L.B.; Arias, C.A. Enterococci: From Commensals to Leading Causes of Drug Resistant Infection. In Enterococcal Infection—Treatment and Antibiotic Resistance, 1st ed.; Gilmore, M.S., Clewell, D.B., Ike, Y., Shankar, N., Eds.; Eye and Ear Infirmary: Boston, MA, USA, 2014; pp. 1–62. Available online: https://www.ncbi.nlm.nih.gov/books/NBK190420/ (accessed on 29 October 2021).
- EUCAST. European Committee on Antimicrobial Susceptibility Testing. Available online: https://www.eucast.org/ast_of_bacteria/ (accessed on 29 October 2021).
- Schito, A.M.; Piatti, G.; Caviglia, D.; Zuccari, G.; Zorzoli, A.; Marimpietri, D.; Alfei, S. Bactericidal Activity of Non-Cytotoxic Cationic Nanoparticles against Clinically and Environmentally Relevant Pseudomonas spp. Isolates. Pharmaceutics 2021, 13, 1411. [Google Scholar] [CrossRef]
- Schito, A.M.; Piatti, G.; Stauder, M.; Bisio, A.; Giacomelli, E.; Romussi, G.; Pruzzo, C. Effects of demethylfruticuline A and fruticuline A from Salvia corrugata Vahl. on biofilm production in vitro by multiresistant strains of Staphylococcus aureus, Staphylococcus epidermidis and Enterococcus faecalis. Int. J. Antimicrob. Agents 2011, 37, 129–134. [Google Scholar] [CrossRef] [Green Version]
- Di Paolo, D.; Pastorino, F.; Zuccari, G.; Caffa, I.; Loi, M.; Marimpietri, D.; Brignole, C.; Perri, P.; Cilli, M.; Nico, B.; et al. Enhanced Anti-Tumor and Anti-Angiogenic Efficacy of a Novel Liposomal Fenretinide on Human Neuroblastoma. J. Control. Release 2013, 170, 445–451. [Google Scholar] [CrossRef]
- Wolska, K.I.; Grudniak, A.M.; Fiecek, B.; Kraczkiewicz-Dowjat, A.; Kurek, A. Antibacterial activity of oleanolic and ursolic acids and their derivatives. Cent. Eur. J. Biol. 2010, 5, 543–553. [Google Scholar] [CrossRef]
- Do Nascimento, P.G.; Lemos, T.L.; Bizerra, A.M.; Arriaga, Â.M.; Ferreira, D.A.; Santiago, G.M.; Braz-Filho, R.; Costa, J.G.M. Antibacterial and Antioxidant Activities of Ursolic Acid and Derivatives. Molecules 2014, 19, 1317–1327. [Google Scholar] [CrossRef]
- Schito, A.M.; Schito, G.C.; Alfei, S. Synthesis and Antibacterial Activity of Cationic Amino Acid-Conjugated Dendrimers Loaded with a Mixture of Two Triterpenoid Acids. Polymers 2021, 13, 521. [Google Scholar] [CrossRef]
- Kurek, A.; Grudniak, A.M.; Szwed, M. Oleanolic acid and ursolic acid affect peptidoglycan metabolism in Listeria monocytogenes. Antonie van Leeuwenhoek 2010, 97, 61. [Google Scholar] [CrossRef]
- Alvarado, H.L.; Abrego, G.; Garduño-Ramirez, M.L.; Clares, B.; Calpena, A.C.; García, M.L. Design and optimization of oleanolic/ursolic acid-loaded nanoplatforms for ocular anti-inflammatory applications. Nanomed. Nanotechnol. Biol. Med. 2015, 11, 521–530. [Google Scholar] [CrossRef]
- Munish, G.; Parul, G. Encapsulation of bio-active compound ursolic acid as proniosomes and its evaluation. Asian J. Pharm. 2013, 7, 158–162. [Google Scholar]
- Antonio, E. Chitosan Modified Poly (Lactic Acid) Nanoparticles Increased the Ursolic Acid Oral Bioavailability. Int. J. Biol. Macromol. 2021, 172, 133–142. [Google Scholar] [CrossRef]
- Wang, L.; Yin, Q.; Liu, C.; Tang, Y.; Sun, C.; Zhuang, J. Nanoformulations of Ursolic Acid: A Modern Natural Anticancer Molecule. Front. Pharmacol. 2021, 12, 1682. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zhao, T.; Liu, Y.; Wang, Q.; Xing, S.; Li, L.; Wang, L.; Liu, L.; Gao, D. Ursolic Acid Liposomes with Chitosan Modification: Promising Antitumor Drug Delivery and Efficacy. Mater. Sci. Eng. C Mater. Biol. Appl. 2017, 71, 1231–1240. [Google Scholar] [CrossRef]
- Jin, H.; Pi, J.; Yang, F.; Jiang, J.; Wang, X.; Bai, H.; Shao, M.; Huang, L.; Zhu, H.; Yang, P.; et al. Folate-Chitosan Nanoparticles Loaded with Ursolic Acid Confer Anti-Breast Cancer Activities in vitro and in vivo. Sci. Rep. 2016, 6, 30782. [Google Scholar] [CrossRef]
- Marena, G.D.; Fonseca-Santos, B.; Matheus Aparecido dos Santos, R.; dos Santos, K.C.; Bauab, T.M.; Chorilli, M. Incorporation of Ursolic Acid in Liquid Crystalline Systems Improves the Antifungal Activity Against Candida Sp. J. Pharm. Innov. 2020. [Google Scholar] [CrossRef]
- Ubeda, C.; Taur, Y.; Jeng, R.R.; Equinda, M.J.; Son, T.; Samstein, M.; Viale, A.; Socci, N.D.; Van Den Brink, M.R.; Kamboj, M.; et al. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J. Clin. Investig. 2010, 120, 4332–4341. [Google Scholar] [CrossRef] [PubMed]
- Arias, C.A.; Contreras, G.A.; Murray, B.E. Emergence and management of drug-resistant enterococcal infections. Expert Rev. Anti-Infect. Ther. 2008, 6, 637–655. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.V.; Weinstock, G.M.; Murray, B.E. An Enterococcus faecalis ABC homologue (Lsa) is required for the resistance of this species to clindamycin and quinupristin-dalfopristin. Antimicrob. Agents Chemother. 2002, 46, 1845–1850. [Google Scholar] [CrossRef] [Green Version]
- Mir, M.; Asong, J.; Li, X.; Cardot, J.; Boons, G.J.; Husson, R.N. The extracytoplasmic domain of the Mycobacterium tuberculosis Ser/Thr kinase PknB binds specific muropeptides and is required for PknB localization. PLoS Pathog. 2011, 7, e1002182. [Google Scholar] [CrossRef] [Green Version]
- Hall, A.D.; Steed, M.E.; Arias, C.A.; Murray, B.E.; Rybak, M.J. Evaluation of Standard- and High-Dose Daptomycin versus Linezolid against Vancomycin-Resistant Enterococcus Isolates in an In Vitro Pharmacokinetic/Pharmacodynamic Model with Simulated Endocardial Vegetations. Antimicrob. Agents Chemother. 2012, 56, 3174–3180. [Google Scholar] [CrossRef] [Green Version]
- Zhanel, G.G.; Laing, N.M.; Nichol, K.A.; Palatnick, L.P.; Noreddin, A.; Hisanaga, T.; Johnson, J.L.; Hoban, D.J. the NAVRESS Group. Antibiotic activity against urinary tract infection (UTI) isolates of vancomycin-resistant enterococci (VRE): Results from the 2002 North American Vancomycin Resistant Enterococci Susceptibility Study (NAVRESS). J. Antimicrob. Chemother. 2003, 52, 382–388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Louis, B.; Rice, L.B. Emergence of Vancomycin-Resistant Enterococci. Emerg. Infect. Dis. 2001, 7, 1–7. [Google Scholar]
- Horiuchi, K.; Shiota, S.; Hatano, T.; Yoshida, T.; Kuroda, T.; Tsuchiya, T. Antimicrobial activity of oleanolic acid from Salvia officinalis and related compounds on vancomycin-resistant enterococci (VRE). Biol. Pharm. Bull. 2007, 30, 1147–1149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schito, A.M.; Alfei, S. Antibacterial Activity of Non-Cytotoxic, Amino Acid-Modified Polycationic Dendrimers against Pseudomonas aeruginosa and Other Non-Fermenting Gram-Negative Bacteria. Polymers 2020, 12, 1818. [Google Scholar] [CrossRef]
- Alfei, S.; Caviglia, D.; Piatti, G.; Zuccari, G.; Schito, A.M. Bactericidal Activity of a Self-Biodegradable Lysine-Containing Dendrimer against Clinical Isolates of Acinetobacter Genus. Int. J. Mol. Sci. 2021, 22, 7274. [Google Scholar] [CrossRef] [PubMed]
- Hyldgaard, M.; Meyer, R.L.; Peng, M.; Hibberd, A.A.; Fischer, J.; Sigmundsson, A.; Mygind, T. Binary combination of epsilon-poly-l-lysine and isoeugenol affect progression of spoilage microbiota in fresh turkey meat, and delay onset of spoilage in Pseudomonas putida challenged meat. Int. J. Food Microbiol. 2015, 215, 131–142. [Google Scholar] [CrossRef]
- Peña-Morán, O.A.; Villarreal, M.L.; Álvarez-Berber, L.; Meneses-Acosta, A.; Rodríguez-López, V. Cytotoxicity, Post-Treatment Recovery, and Selectivity Analysis of Naturally Occurring Podophyllotoxins from Bursera Fagaroides Var. Fagaroides on Breast Cancer Cell Lines. Molecules 2016, 21, 1013. [Google Scholar] [CrossRef]
- Awouafack, M.D.; McGaw, L.J.; Gottfried, S.; Mbouangouere, R.; Tane, P.; Spiteller, M.; Eloff, J.N. Antimicrobial Activity and Cytotoxicity of the Ethanol Extract, Fractions and Eight Compounds Isolated from Eriosema Robustum (Fabaceae). BMC Complementary Altern. Med. 2013, 13, 289. [Google Scholar] [CrossRef] [Green Version]
- Weerapreeyakul, N.; Nonpunya, A.; Barusrux, S.; Thitimetharoch, T.; Sripanidkulchai, B. Evaluation of the Anticancer Potential of Six Herbs against a Hepatoma Cell Line. Chin. Med. 2012, 7, 15. [Google Scholar] [CrossRef] [Green Version]
- Adamu, M.; Naidoo, V.; Eloff, J.N. The Antibacterial Activity, Antioxidant Activity and Selectivity Index of Leaf Extracts of Thirteen South African Tree Species Used in Ethnoveterinary Medicine to Treat Helminth Infections. BMC Vet. Res. 2014, 10, 52. [Google Scholar] [CrossRef] [Green Version]
- Adamu, M.; Naidoo, V.; Eloff, J.N. Efficacy and Toxicity of Thirteen Plant Leaf Acetone Extracts Used in Ethnoveterinary Medicine in South Africa on Egg Hatching and Larval Development of Haemonchus Contortus. BMC Vet. Res. 2013, 9, 38. [Google Scholar] [CrossRef] [Green Version]
- Famuyide, I.M.; Aro, A.O.; Fasina, F.O.; Eloff, J.N.; McGaw, L.J. Antibacterial and Antibiofilm Activity of Acetone Leaf Extracts of Nine Under-Investigated South African Eugenia and Syzygium (Myrtaceae) Species and Their Selectivity Indices. BMC Complementary Altern. Med. 2019, 19, 141. [Google Scholar] [CrossRef] [Green Version]
- Nogueira, F.; do Rosario, V.E. Methods for assessment of antimalarial activity in the different phases of the Plasmodiumlife cycle. Rev. Pan-Amaz. Saude 2010, 1, 109–124. [Google Scholar] [CrossRef]






| Analysis | UA-G4K | |
|---|---|---|
| FTIR | NH3+ (dendrimer) | 3500–3000 |
| (cm−1) | OH stretching (UA) | 3500–2500 |
| –C=O esters (dendrimer) | 1735 | |
| –C=O carboxyl (UA) | 1688 | |
| –C–O esters (dendrimer) | 1215, 1244 | |
| 1H NMR ° | CH3 + H (C (5)) of UA, several s, 726H | 0.75–0.98 |
| (400 MHz, CD3OD) | CH3 G1–G4 + CH2CH2CH2 lys + CH2 UA + CH UA, m, 1116H | 1.00–2.40 |
| (ppm) | CH2NH3+ Lys + CH UA, m, 129H | 2.95–3.16 |
| CH2O dendrimer + CHNH3+ lys, m, 234H | 4.10–4.30 | |
| CH UA, m, 33H | 4.58–4.70 | |
| CH UA, m, 33H | 5.22 | |
| Elemental Analysis | C, H, N, Cl | 60.64, 8.49, 4.96, 11.00 * |
| 60.24, 8.81, 4.77, 11.32 § | ||
| HPLC | Retention Time (min) | 19.86 |
| DL (%) | 49.7 ± 5.9 | |
| EE (%) | 59.1 ± 5.9 | |
| 1H NMR | MW | 30,069 |
| DL% by HPLC | 29,804.5 ± 1735.5 | |
| DLS 1 Analysis | Z-Ave 2 (nm) | 577.5 ± 10.7 2,5 |
| PDI 3 | 0.235 ± 0.028 3,5 | |
| Z-potential 4 (ζ-p) | −42.6 ± 4.4 4,5 | |
| Solubilization Essay | Water Solubility (mg/mL) | 10.2 |
| Dialysis Method (HPLC) | Cumulative Release (%, 24 h) | 55.7 |
| Mathematical Model | Higuchi (Q = KHt1/2)6 | |
| Mechanism | Diffusion based on Ficks’ Law | |
| Cytotoxicity | Cell Viability (%) | 94.3 8 |
| (HeLa Cells) | (20 µM) 7 | |
| MIC Values 1 | UA | UA-G4K | Max UA Concentration 2 |
|---|---|---|---|
| MW 456.7 | MW 30,069 | ||
| µg/mL, µM | µg/mL, µM | µg/mL, µM | |
| Gram-Negative Enterobacteriaceae and Non-Fermenting Bacteria | |||
| E. coli 224 S # | >128, >280.3 | >128, >4.3 | >35.4, >77.6 |
| P. aeruginosa 247 | >128, >280.3 | >128, >4.3 | >35.4, >77.6 |
| Gram-Positive Staphylococci | |||
| S. aureus 18 * | 16–32, >35–70 | >128, >4.3 | >35.4, >77.6 |
| S. epidermidis 22 † | 16–32, >35–70 | >128, >4.3 | >35.4, >77.6 |
| MIC Values 1 | UA | UA-G4K | Max UA Released 2 | Reference Antibiotic | SI 3 UA | SI 3 |
|---|---|---|---|---|---|---|
| MW 456.7 | MW 30,069 | UA-G4K | ||||
| µg/mL, µM | µg/mL, µM | µg/mL, µM | µg/mL, µM | |||
| E. faecalis 1 * | 2, 4.4 | 64, 2.1 | 17.7, 38.8 | 128 (366.3) 4 | 12.5 | 46.1 |
| 512 (353.3) 5 | ||||||
| E. faecalis 4 # | 2, 4.4 | 128, 4.3 | 35.4, 77.6 | 0.5–4 (1.4–11.4) 4 | 12.5 | 22.5 |
| 2.0 (1.4) 5 | ||||||
| E. faecalis 8 # | 2, 4.4 | 128, 4.3 | 35.4, 77.6 | 0.5–4 (1.4–11.4) 4 | 12.5 | 22.5 |
| 2.0 (1.4) 5 | ||||||
| E. faecalis 79 * | 4, 8.8 | 64, 2.1 | 17.7, 38.8 | 128 (366.3) 4 | 6.2 | 46.1 |
| 512 (353.3) 5 | ||||||
| E. faecalis 108 # | 2, 4.4 | 32, 1.1 | 8.9, 19.4 | 0.5–4 (1.4–11.4) 4 | 12.5 | 88.1 |
| 2.0 (1.4) 5 | ||||||
| E. faecalis 200 # | 2, 4.4 | 32, 1.1 | 8.9, 19.4 | 0.5–4 (1.4–11.4) 4 | 12.5 | 88.1 |
| 2.0 (1.4) 5 | ||||||
| E. faecalis 261 # | 4, 8.8 | 128, 4.3 | 17.7, 38.8 | 0.5–4 (1.4–11.4) 4 | 6.2 | 22.5 |
| 2.0 (1.4) 5 | ||||||
| E. faecalis 425 # | 4, 8.8 | 64, 2.1 | 17.7, 38.8 | 0.5–4 (1.4–11.4) 4 | 6.2 | 46.1 |
| 2.0 (1.4) 5 | ||||||
| E. faecalis 19 †,* | 2, 4.4 | 128, 4.3 | 35.4, 77.6 | 128 (366.3) 4 | 12.5 | 22.5 |
| 512 (353.3) 5 | ||||||
| E. faecalis 51 †,* | 2, 4.4 | 64, 2.1 | 17.7, 38.8 | 128 (366.3) 4 | 12.5 | 46.1 |
| 512 (353.3) 5 | ||||||
| E. faecium 21 # | 2, 4.4 | 64, 2.1 | 17.7, 38.8 | 0.5–4 (1.4–11.4) 4 | 12.5 | 46.1 |
| 2.0 (1.4) 5 | ||||||
| E. faecium 43 * | 2, 4.4 | 32, 1.1 | 8.9, 19.4 | 128 (366.3) 4 | 12.5 | 88.1 |
| 512 (353.3) 5 | ||||||
| E. faecium 152 * | 2, 4.4 | 32, 1.1 | 8.9, 19.4 | 128 (366.3) 4 | 12.5 | 88.1 |
| 512 (353.3) 5 | ||||||
| E. faecium 185 * | 4, 8.8 | 32, 1.1 | 8.9, 19.4 | 128 (366.3) 4 | 6.2 | 88.1 |
| 512 (353.3) 5 | ||||||
| E. faecium 341 * | 2, 4.4 | 64, 2.1 | 17.7, 38.8 | 128 (366.3) 4 | 12.5 | 46.1 |
| 512 (353.3) 5 | ||||||
| E. faecium 3 †,* | 2, 4.4 | 32, 1.1 | 8.9, 19.4 | 128 (366.3) 4 | 12.5 | 88.1 |
| 512 (353.3) 5 | ||||||
| E. gallinarum 150 | 2, 4.4 | 32, 1.1 | 8.9, 19.4 | N.R. | 12.5 | 88.1 |
| E. gallinarum 294 | 2, 4.4 | 16, 0.5 | 4.5, 9.7 | N.R. | 12.5 | 193.8 |
| E. gallinarum 295 | 4, 8.8 | 16, 0.5 | 4.5, 9.7 | N.R. | 6.2 | 193.8 |
| E. casseliflavus 184 | 4, 8.8 | 32, 1.1 | 8.9, 19.4 | N.R. | 6.2 | 88.1 |
| E. casseliflavus 296 | 2, 4.4 | 32, 1.1 | 8.9, 19.4 | N.R. | 12.5 | 88.1 |
| E. casseliflavus 159 | 2, 4.4 | 16, 0.5 | 4.5, 9.7 | N.R. | 12.5 | 193.8 |
| E. avium 119 | 2, 4.4 | 16, 0.5 | 4.5, 9.7 | N.R. | 12.5 | 193.8 |
| E. avium 122 | 2, 4.4 | 16, 0.5 | 4.5, 9.7 | N.R. | 12.5 | 193.8 |
| E. avium 167 | 4, 8.8 | 32, 1.1 | 8.9, 19.4 | N.R. | 6.2 | 88.1 |
| E. durans 103 | 2, 4.4 | 64, 2.1 | 17.7, 38.8 | N.R. | 12.5 | 46.1 |
| E. durans 113 | 4, 8.8 | 64, 2.1 | 17.7, 38.8 | N.R. | 6.2 | 46.1 |
| Sample | Equations | R2 | LD50 (µM) | SI |
|---|---|---|---|---|
| G4K | y = −0.7300x + 84.601 | 0.9000 | 47.4 | N.D |
| UA | y = −0.8206x + 95.040 | 0.9606 | 54.9 | 6.2–12.5 |
| UA-G4K | y = −0.4386x + 92.482 | 0.9202 | 96.9 | 22.5–193.7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schito, A.M.; Caviglia, D.; Piatti, G.; Zorzoli, A.; Marimpietri, D.; Zuccari, G.; Schito, G.C.; Alfei, S. Efficacy of Ursolic Acid-Enriched Water-Soluble and Not Cytotoxic Nanoparticles against Enterococci. Pharmaceutics 2021, 13, 1976. https://doi.org/10.3390/pharmaceutics13111976
Schito AM, Caviglia D, Piatti G, Zorzoli A, Marimpietri D, Zuccari G, Schito GC, Alfei S. Efficacy of Ursolic Acid-Enriched Water-Soluble and Not Cytotoxic Nanoparticles against Enterococci. Pharmaceutics. 2021; 13(11):1976. https://doi.org/10.3390/pharmaceutics13111976
Chicago/Turabian StyleSchito, Anna Maria, Debora Caviglia, Gabriella Piatti, Alessia Zorzoli, Danilo Marimpietri, Guendalina Zuccari, Gian Carlo Schito, and Silvana Alfei. 2021. "Efficacy of Ursolic Acid-Enriched Water-Soluble and Not Cytotoxic Nanoparticles against Enterococci" Pharmaceutics 13, no. 11: 1976. https://doi.org/10.3390/pharmaceutics13111976
APA StyleSchito, A. M., Caviglia, D., Piatti, G., Zorzoli, A., Marimpietri, D., Zuccari, G., Schito, G. C., & Alfei, S. (2021). Efficacy of Ursolic Acid-Enriched Water-Soluble and Not Cytotoxic Nanoparticles against Enterococci. Pharmaceutics, 13(11), 1976. https://doi.org/10.3390/pharmaceutics13111976