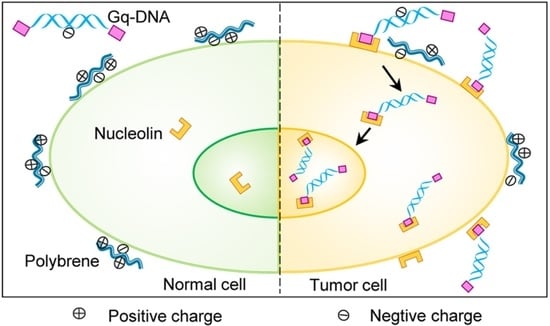
G-Quadruplex Linked DNA Guides Selective Transfection into Nucleolin-Overexpressing Cancer Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. DNA Linearization
2.2. Amplification of Single Strand DNA
2.3. Generation and Purification of Gq-DNA
2.4. Cells and Cell Culture
2.5. Serum Stability Assay
2.6. DNA Delivery
2.7. Fluorescent Staining
2.8. Flow Cytometry
2.9. Luciferase Assay
2.10. Cellular Uptake
2.11. Quantitative Real-Time PCR Analysis
2.12. MTT Assay
2.13. Lactic Dehydrogenase (LDH) Leakage Assay
2.14. RNA Interference
2.15. Statistics Analysis
3. Results
3.1. Synthetic Procedure of Gq-DNA
3.2. Characterization of Gq-DNA
3.3. Gq-DNA Transfects Tumor Cells after Neutralizing Negative Charges of Cell Membrane
3.4. Gq-DNA Transfection Effectively Targets Tumor Cells with High-Level Nucleolin
3.5. Gq-DNA Transfection Works Independently of Cell Proliferation
3.6. Gq-DNA Transfection Depends on the Binding between G-Quadruplex and Nucleolin
3.7. Gq Facilitates the Entrance of Gq-DNA into Cells and Nuclei
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farmer, Z.L.; Kim, E.S.; Carrizosa, D.R. Gene Therapy in Head and Neck Cancer. Oral. Maxillofac. Surg. Clin. N. Am. 2019, 31, 117–124. [Google Scholar] [CrossRef] [PubMed]
- High, K.A.; Roncarolo, M.G. Gene Therapy. N. Engl. J. Med. 2019, 381, 455–464. [Google Scholar] [CrossRef] [PubMed]
- Dunbar, C.E.; High, K.A.; Joung, J.K.; Kohn, D.B.; Ozawa, K.; Sadelain, M. Gene therapy comes of age. Science 2018, 359, eaan4672. [Google Scholar] [CrossRef] [Green Version]
- van Haasteren, J.; Li, J.; Scheideler, O.J.; Murthy, N.; Schaffer, D.V. The delivery challenge: Fulfilling the promise of therapeutic genome editing. Nat. Biotechnol. 2020, 38, 845–855. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.; Arora, S.; Singh, J.; Layek, B. A review of the tortuous path of nonviral gene delivery and recent progress. Int. J. Biol. Macromol. 2021, 183, 2055–2073. [Google Scholar] [CrossRef]
- Uludag, H.; Ubeda, A.; Ansari, A. At the Intersection of Biomaterials and Gene Therapy: Progress in Non-viral Delivery of Nucleic Acids. Front. Bioeng. Biotechnol. 2019, 7, 131. [Google Scholar] [CrossRef]
- Zhi, D.; Bai, Y.; Yang, J.; Cui, S.; Zhao, Y.; Chen, H.; Zhang, S. A review on cationic lipids with different linkers for gene delivery. Adv. Colloid Interface Sci. 2018, 253, 117–140. [Google Scholar] [CrossRef]
- Nakase, M.; Okumura, K.; Kamei, T.; Nakamura, S.; Inui, M.; Tagawa, T. The influence of oral tumor cell proliferation activity and membrane potential on the transfection efficiency of a cationic liposome. Oncol. Rep. 2005, 14, 1487–1491. [Google Scholar] [CrossRef] [Green Version]
- Mannisto, M.; Ronkko, S.; Matto, M.; Honkakoski, P.; Hyttinen, M.; Pelkonen, J.; Urtti, A. The role of cell cycle on polyplex-mediated gene transfer into a retinal pigment epithelial cell line. J. Gene Med. 2005, 7, 466–476. [Google Scholar] [CrossRef]
- Romano, S.; Fonseca, N.; Simoes, S.; Goncalves, J.; Moreira, J.N. Nucleolin-based targeting strategies for cancer therapy: From targeted drug delivery to cytotoxic ligands. Drug Discov. Today 2019, 24, 1985–2001. [Google Scholar] [CrossRef] [PubMed]
- Ugrinova, I.; Petrova, M.; Chalabi-Dchar, M.; Bouvet, P. Multifaceted Nucleolin Protein and Its Molecular Partners in Oncogenesis. Adv. Protein Chem. Struct. Biol. 2018, 111, 133–164. [Google Scholar]
- Gonzalez, V.; Guo, K.; Hurley, L.; Sun, D. Identification and characterization of nucleolin as a c-myc G-quadruplex-binding protein. J. Biol. Chem. 2009, 284, 23622–23635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varshney, D.; Spiegel, J.; Zyner, K.; Tannahill, D.; Balasubramanian, S. The regulation and functions of DNA and RNA G-quadruplexes. Nat. Rev. Mol. Cell Biol. 2020, 21, 459–474. [Google Scholar] [CrossRef] [PubMed]
- Harkness, R.W.T.; Mittermaier, A.K. G-quadruplex dynamics. Biochim. Biophys Acta Proteins Proteom 2017, 1865 (11 Pt B), 1544–1554. [Google Scholar] [CrossRef]
- Teng, F.Y.; Jiang, Z.Z.; Guo, M.; Tan, X.Z.; Chen, F.; Xi, X.G.; Xu, Y. G-quadruplex DNA: A novel target for drug design. Cell. Mol. Life Sci 2021, 78, 6557–6583. [Google Scholar] [CrossRef]
- Puig Lombardi, E.; Holmes, A.; Verga, D.; Teulade-Fichou, M.P.; Nicolas, A.; Londono-Vallejo, A. Thermodynamically stable and genetically unstable G-quadruplexes are depleted in genomes across species. Nucleic Acids Res. 2019, 47, 6098–6113. [Google Scholar] [CrossRef] [Green Version]
- Kwok, C.K.; Merrick, C.J. G-Quadruplexes: Prediction, Characterization, and Biological Application. Trends Biotechnol. 2017, 35, 997–1013. [Google Scholar] [CrossRef]
- Monsen, R.C.; Trent, J.O. G-quadruplex virtual drug screening: A review. Biochimie 2018, 152, 134–148. [Google Scholar] [CrossRef]
- Dailey, M.M.; Miller, M.C.; Bates, P.J.; Lane, A.N.; Trent, J.O. Resolution and characterization of the structural polymorphism of a single quadruplex-forming sequence. Nucleic Acids Res. 2010, 38, 4877–4888. [Google Scholar] [CrossRef]
- Reyes-Reyes, E.M.; Teng, Y.; Bates, P.J. A new paradigm for aptamer therapeutic AS1411 action: Uptake by macropinocytosis and its stimulation by a nucleolin-dependent mechanism. Cancer Res. 2010, 70, 8617–8629. [Google Scholar] [CrossRef]
- Yazdian-Robati, R.; Bayat, P.; Oroojalian, F.; Zargari, M.; Ramezani, M.; Taghdisi, S.M.; Abnous, K. Therapeutic applications of AS1411 aptamer, an update review. Int. J. Biol. Macromol. 2020, 155, 1420–1431. [Google Scholar] [CrossRef] [PubMed]
- Dapic, V.; Bates, P.J.; Trent, J.O.; Rodger, A.; Thomas, S.D.; Miller, D.M. Antiproliferative activity of G-quartet-forming oligonucleotides with backbone and sugar modifications. Biochemistry 2002, 41, 3676–3685. [Google Scholar] [CrossRef]
- Nasri, M.; Karimi, A.; Allahbakhshian Farsani, M. Production, purification and titration of a lentivirus-based vector for gene delivery purposes. Cytotechnology 2014, 66, 1031–1038. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacobsen, F.; Hirsch, T.; Mittler, D.; Schulte, M.; Lehnhardt, M.; Druecke, D.; Homann, H.H.; Steinau, H.U.; Steinstraesser, L. Polybrene improves transfection efficacy of recombinant replication-deficient adenovirus in cutaneous cells and burned skin. J. Gene Med. 2006, 8, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Wu, N.; Deng, F.; Zhang, H.; Wang, N.; Zhang, W.; Chen, X.; Wen, S.; Zhang, J.; Yin, L.; et al. Adenovirus-mediated gene transfer in mesenchymal stem cells can be significantly enhanced by the cationic polymer polybrene. PLoS ONE 2014, 9, e92908. [Google Scholar] [CrossRef] [PubMed]
- Chaney, W.G.; Howard, D.R.; Pollard, J.W.; Sallustio, S.; Stanley, P. High-frequency transfection of CHO cells using polybrene. Somat. Cell Mol. Genet. 1986, 12, 237–244. [Google Scholar] [CrossRef] [PubMed]
- Morgan, T.L.; Maher, V.M.; McCormick, J.J. Optimal parameters for the polybrene-induced DNA transfection of diploid human fibroblasts. Vitr. Cell. Dev. Biol. 1986, 22, 317–319. [Google Scholar] [CrossRef] [PubMed]
- Chaney, W.G.; Howard, D.R.; Pollard, J.W.; Sallustio, S.; Stanley, P.; Walker, J.M.; Chaney, W.G. DNA transfection of Mammalian cells using polybrene. In New Nucleic Acid Techniques; Humana Press: Totowa, NJ, USA, 1988; Volume 4, pp. 363–370. [Google Scholar]
- Gigante, A.; Li, M.; Junghänel, S.; Hirschhäuser, C.; Knauer, S.; Schmuck, C. Non-viral transfection vectors: Are hybrid materials the way forward? MedChemComm 2019, 10, 1692–1718. [Google Scholar] [CrossRef]
- Hovanessian, A.G.; Soundaramourty, C.; El Khoury, D.; Nondier, I.; Svab, J.; Krust, B. Surface expressed nucleolin is constantly induced in tumor cells to mediate calcium-dependent ligand internalization. PLoS ONE 2010, 5, e15787. [Google Scholar] [CrossRef] [Green Version]
- Qin, R.; Zhang, H.; Li, S.; Jiang, W.; Liu, D. Three major nucleolar proteins migrate from nucleolus to nucleoplasm and cytoplasm in root tip cells of Vicia faba L. exposed to aluminum. Environ. Sci. Pollut. Res. Int. 2014, 21, 10736–10743. [Google Scholar] [CrossRef] [PubMed]
- Ferrara, B.; Belbekhouche, S.; Habert, D.; Houppe, C.; Vallee, B.; Bourgoin-Voillard, S.; Cohen, J.L.; Cascone, I.; Courty, J. Cell surface nucleolin as active bait for nanomedicine in cancer therapy: A promising option. Nanotechnology 2021, 32, 322001–322027. [Google Scholar] [CrossRef] [PubMed]
- Graessmann, M.; Menne, J.; Liebler, M.; Graeber, I. Helper activity for gene expression, a novel function of the SV40 enhancer. Nucleic Acids Res. 1989, 17, 6603–6612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.; Shi, H.; Zhou, H.; Song, X.; Yuan, S.; Luo, Y. The angiogenic function of nucleolin is mediated by vascular endothelial growth factor and nonmuscle myosin. Blood 2006, 107, 3564–3571. [Google Scholar] [CrossRef] [PubMed]
- Viallard, C.; Larrivee, B. Tumor angiogenesis and vascular normalization: Alternative therapeutic targets. Angiogenesis 2017, 20, 409–426. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, J.; Alway, S.E. Transgene expression in hypertrophied and aged skeletal muscle in vivo by lentivirus delivery. J. Gene Med. 2004, 6, 278–287. [Google Scholar] [CrossRef] [PubMed]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiang, M.; Li, Y.; Liu, J.; Shi, J.; Ge, Y.; Peng, C.; Bin, Y.; Wang, Z.; Wang, L. G-Quadruplex Linked DNA Guides Selective Transfection into Nucleolin-Overexpressing Cancer Cells. Pharmaceutics 2022, 14, 2247. https://doi.org/10.3390/pharmaceutics14102247
Xiang M, Li Y, Liu J, Shi J, Ge Y, Peng C, Bin Y, Wang Z, Wang L. G-Quadruplex Linked DNA Guides Selective Transfection into Nucleolin-Overexpressing Cancer Cells. Pharmaceutics. 2022; 14(10):2247. https://doi.org/10.3390/pharmaceutics14102247
Chicago/Turabian StyleXiang, Mengxi, Yongkui Li, Jia Liu, Jie Shi, Yizhi Ge, Chen Peng, Yawen Bin, Zheng Wang, and Lin Wang. 2022. "G-Quadruplex Linked DNA Guides Selective Transfection into Nucleolin-Overexpressing Cancer Cells" Pharmaceutics 14, no. 10: 2247. https://doi.org/10.3390/pharmaceutics14102247
APA StyleXiang, M., Li, Y., Liu, J., Shi, J., Ge, Y., Peng, C., Bin, Y., Wang, Z., & Wang, L. (2022). G-Quadruplex Linked DNA Guides Selective Transfection into Nucleolin-Overexpressing Cancer Cells. Pharmaceutics, 14(10), 2247. https://doi.org/10.3390/pharmaceutics14102247