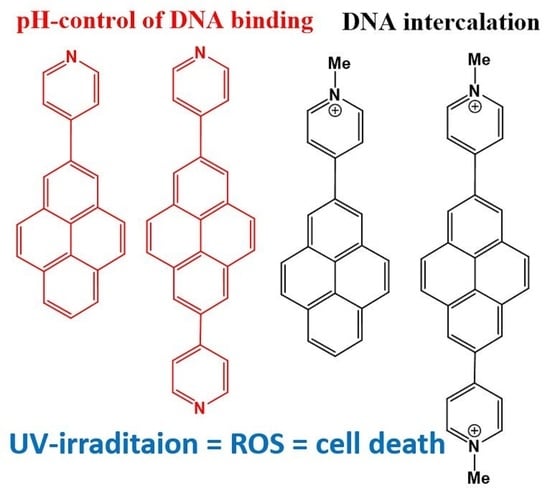
Para-N-Methylpyridinium Pyrenes: Impact of Positive Charge on ds-DNA/RNA and Protein Recognition, Photo-Induced Bioactivity, and Intracellular Localisation
Abstract
:1. Introduction
2. Materials and Methods
Study of Interactions with ds-DNA and ds-RNA
3. Results
3.1. Protonation Properties of Neutral Analogues 1 and 2
3.2. Non-Covalent Interactions of 1, 2, 1M and 2M with Various ds-DNA, ds-RNA, and BSA
3.2.1. Thermal Denaturation of ds-DNA/RNA
3.2.2. Spectrophotometric Titrations of Pyrene Analogues with DNA, RNA, BSA
3.2.3. Circular Dichroism (CD) Experiments
3.2.4. 2M and 1M Induce Plasmid DNA Cleavage
4. Biological Activity of 1, 2, 1M, and 2M
4.1. UV Light Irradiation Increases the Cell Toxicity of 2M, 1 or 1M
4.2. UV Light Irradiation Increases ROS Production in Cells Exposed to 2M, 1, or 1M
4.3. UV Light Irradiation Increases Cell Size of 2M, 1 or 1M Treated A549 Cells
4.4. Intracellular Localisation of Compounds
4.5. 1M in Combination with UV Light Irradiation Acts on Mitochondria Causing Their Dysfunction by Increasing Their ROS Levels
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Aerssens, D.; Cadoni, E.; Tack, L.; Madder, A. Photosensitized Singlet Oxygen (1O2) Toolbox for Bio-Organic Applications: Tailoring 1O2 Generation for DNA and Protein Labelling, Targeting and Biosensing. Molecules 2022, 27, 778. [Google Scholar] [CrossRef] [PubMed]
- Dolmans, D.E.; Fukumura, D.; Jain, R.K. Photodynamic therapy for cancer. Nat. Rev. Cancer 2003, 3, 380–387. [Google Scholar] [CrossRef] [PubMed]
- Danielson, B.G. Structure, chemistry, and pharmacokinetics of intravenous iron agents. J. Am. Soc. Nephrol. 2004, 15, S93–S98. [Google Scholar] [PubMed]
- Chisholm, D.R.; Lamb, R.; Pallett, T.; Affleck, V.; Holden, C.; Marrison, J.; O’Toole, P.; Ashton, P.D.; Newling, K.; Steffen, A.; et al. Photoactivated cell-killing involving a low molecular weight, donor-acceptor diphenylacetylene. Chem. Sci. 2019, 10, 4673–4683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abrahamse, H.; Hamblin, M.R. New photosensitizers for photodynamic therapy. Biochem. J. 2016, 473, 347–364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banik, B.K.; Becker, F.F. Synthesis, electrophilic substitution and structure-activity relationship studies of polycyclic aromatic compounds towards the development of anticancer agents. Curr. Med. Chem. 2001, 8, 1513–1533. [Google Scholar] [CrossRef]
- Kanamori, T.; Matsuyama, A.; Naito, H.; Tsuga, Y.; Ozako, Y.; Ogura, S.; Okazaki, S.; Yuasa, H. Water-Soluble Glucosyl Pyrene Photosensitizers: An Intramolecularly Synthesized 2-C-Glucoside and an O-Glucoside. J. Org. Chem. 2018, 83, 13765–13775. [Google Scholar] [CrossRef]
- Chowdhury, N.; Dutta, S.; Dasgupta, S.; Singh, N.D.P.; Baidya, M.; Ghosh, S.K. Synthesis, photophysical, photochemical, DNA cleavage/binding and cytotoxic properties of pyrene oxime ester conjugates. Photochem. Photobiol. Sci. 2012, 11, 1239–1250. [Google Scholar] [CrossRef]
- Ma, J.-K.; Fathy, W.; Eldin, S.; Rizk El-Ghareeb, W.; Elhelaly, A.E.; Khedr, M.H.E.; Li, X.; Huang, X.-C. Effects of Pyrene on Human Liver HepG2 Cells: Cytotoxicity, Oxidative Stress and Transcriptomic Changes in Xenobiotic Metabolizing Enzymes and Inflammatory Markers with Protection Trial Using Lycopene. BioMed Res. Int. 2019, 2019, 7604851. [Google Scholar] [CrossRef] [Green Version]
- Østergaard, M.E.; Hrdlicka, P.J. Pyrene-functionalized oligonucleotides and locked nucleic acids (LNAs): Tools for fundamental research, diagnostics, and nanotechnology. Chem. Soc. Rev. 2011, 40, 5771–5788. [Google Scholar] [CrossRef]
- Wu, C.; Wang, C.; Yan, L.; Yang, C.J. Pyrene excimer nucleic acid probes for bimolecule signaling. J. Biomed. Nanotechnol. 2009, 5, 495–504. [Google Scholar] [CrossRef] [PubMed]
- Christensen, U.B.; Pedersen, E.B. Intercalating nucleic acids containing insertions of 1-O-(1-pyrenylmethyl)glycerol: Stabilisation of dsDNA and discrimination of DNA over RNA. Nucleic Acids Res. 2002, 30, 4918–4925. [Google Scholar] [CrossRef] [PubMed]
- Filichev, V.V.; Pedersen, E.B. Intercalating nucleic acids (INAs) with insertion of N-(pyren-1-ylmethyl)-(3R,4R)-4-(hydroxymethyl)pyrrolidin-3-ol. DNA (RNA) duplex and DNA three-way junction stabilities. Org. Biomol. Chem. 2003, 1, 100–103. [Google Scholar] [CrossRef] [PubMed]
- Gröger, K.; Baretić, D.; Piantanida, I.; Marjanović, M.; Kralj, M.; Grabar, M.; Tomić, S.; Schmuck, C. Guanidiniocarbonyl-pyrrole-aryl conjugates as nucleic acid sensors: Switch of binding mode and spectroscopic responses by introducing additional binding sites into the linker. Org. Biomol. Chem. 2011, 9, 198–209. [Google Scholar] [CrossRef]
- Astakhova, I.V.; Malakhov, A.D.; Stepanova, I.A.; Ustinov, A.V.; Bondarev, S.L.; Paramonov, A.S.; Korshun, V.A. 1-Phenylethynylpyrene (1-PEPy) as refined excimer forming alternative to pyrene: Case of DNA major groove excimer. Bioconj. Chem. 2007, 18, 1972–1980. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Folgado, L.; Schmuck, C.; Tomić, S.; Piantanida, I. A novel pyrene-guanidiniocarbonyl-pyrrole cation efficiently differentiates between ds-DNA and ds-RNA by two independent, sensitive spectroscopic methods. Bioorg. Med. Chem. Lett. 2008, 18, 2977–2981. [Google Scholar] [CrossRef]
- Hernandez-Folgado, L.; Baretić, D.; Piantanida, I.; Marjanović, M.; Kralj, M.; Rehm, T.; Schmuck, C. Guanidiniocarbonylpyrrole-Aryl Derivatives: Structure Tuning for Spectrophotometric Recognition of Specific DNA and RNA Sequences and for Antiproliferative Activity. Chem. Eur. J. 2010, 16, 3036–3056. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Zou, Y.; Li, C.; Sicking, W.; Piantanida, I.; Yi, T.; Schmuck, C. A Molecular Peptide Beacon for the Ratiometric Sensing of Nucleic Acids. J. Am. Chem. Soc. 2012, 134, 1958–1961. [Google Scholar] [CrossRef]
- Ma, F.; Liu, W.-J.; Qianyi, Z.; Zhang, C.-Y. Sensitive detection of microRNAs by duplex specific nuclease-assisted target recycling and pyrene excimer switching. Chem. Commun. 2017, 53, 10596–10599. [Google Scholar] [CrossRef]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy, 3rd ed.; Springer: Boston MA, USA, 2006. [Google Scholar]
- Ayyavoo, K.; Velusamy, P. Pyrene based materials as fluorescent probes in chemical and biological fields. New J. Chem. 2021, 45, 10997–11017. [Google Scholar] [CrossRef]
- Wanninger-Weiß, C.; Wagenknecht, H.-A. Synthesis of 5-(2-Pyrenyl)-2′-deoxyuridine as a DNA Modification for Electron-Transfer Studies: The Critical Role of the Position of the Chromophore Attachment. Eur. J. Org. Chem. 2008, 2008, 64–71. [Google Scholar] [CrossRef]
- Mayer-Enthart, E.; Wagenknecht, H.-A. Structure-Sensitive and Self-Assembled Helical Pyrene Array Based on DNA Architecture. Angew. Chem. Int. Ed. 2006, 45, 3372–3375. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.T.; Shao, X.D.; Zhao, J.Z.; Wu, M.B. Controllable Photodynamic Therapy Implemented by Regulating Singlet Oxygen Efficiency. Adv. Sci. 2017, 4, 1700113. [Google Scholar] [CrossRef] [PubMed]
- Bolze, F.; Jenni, S.; Sour, A.; Heitz, V. Molecular photosensitisers for two-photon photodynamic therapy. Chem. Commun. 2017, 30, 12857–12877. [Google Scholar] [CrossRef]
- Rumi, M.; Perry, J.W. Two-photon absorption: An overview of measurements and principles. Adv. Opt. Photonics 2010, 2, 451–518. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, L.; Cao, H.Q.; Li, Q.; Li, Y.; Han, M.J.; Wang, H.; Li, J.B. Photodynamic Therapy with Liposomes Encapsulating Photosensitizers with Aggregation-Induced Emission. Nano Lett. 2019, 19, 1821–1826. [Google Scholar] [CrossRef]
- Sun, B.B.; Wang, L.; Li, Q.; He, P.P.; Liu, H.L.; Wang, H.; Yang, Y.; Li, J.B. Bis(pyrene)-Doped Cationic Dipeptide Nanoparticles for Two-Photon-Activated Photodynamic Therapy. Biomacromolecules 2017, 18, 3506–3513. [Google Scholar] [CrossRef]
- Zhang, Y.; Tan, L.; Shia, J.; Ji, L. Iridium-catalysed borylation of pyrene—A powerful route to novel optoelectronic materials. New J. Chem. 2021, 45, 14869–14878. [Google Scholar] [CrossRef]
- Coventry, D.N.; Batsanov, A.S.; Goeta, A.E.; Howard, J.A.; Marder, T.B.; Perutz, R.N. Selective Ir-Catalysed Borylation of Polycyclic Aromatic Hydrocarbons: Structures of Naphthalene-2,6-bis(boronate), Pyrene-2,7-bis(boronate) and Perylene-2,5,8,11-tetra(boronate) Esters. Chem. Commun. 2005, 41, 2172–2174. [Google Scholar] [CrossRef]
- Crawford, A.G.; Dwyer, A.D.; Liu, Z.; Steffen, A.; Beeby, A.; P˚alsson, L.-O.; Tozer, D.J.; Marder, T.B. Experimental and Theoretical Studies of the Photophysical Properties of 2- and 2,7-Functionalized Pyrene Derivatives. J. Am. Chem. Soc. 2011, 133, 13349–13362. [Google Scholar] [CrossRef]
- Crawford, A.G.; Liu, Z.; Mkhalid, I.A.I.; Thibault, M.-H.; Schwarz, N.; Alcaraz, G.; Steffen, A.; Collings, J.C.; Batsanov, A.S.; Howard, J.A.K.; et al. Synthesis of 2- and 2,7-Functionalized Pyrene Derivatives: An Application of Selective C–H Borylation. Chem. Eur. J. 2012, 18, 5022–5035. [Google Scholar] [CrossRef] [PubMed]
- Edkins, R.M.; Fucke, K.; Peach, M.J.G.; Crawford, A.G.; Marder, T.B.; Beeby, A. Synthesis, Structures and Comparison of the Photophysical Properties of Cyclometalated Iridium Complexes Containing the Isomeric 1- and 2-(2′-pyridyl)pyrene Ligands. Inorg. Chem. 2013, 52, 9842–9898. [Google Scholar] [CrossRef] [PubMed]
- Ji, L.; Edkins, R.M.; Lorbach, A.; Krummenacher, I.; Brückner, C.; Eichhorn, A.; Braunschweig, H.; Engels, B.; Low, P.J.; Marder, T.B. Electron Delocalization in Reduced Forms of 2-(BMes2)pyrene and 2,7-Bis(BMes2)pyrene. J. Am. Chem. Soc. 2015, 137, 6750–6753. [Google Scholar] [CrossRef]
- Ji, L.; Lorbach, A.; Edkins, R.M.; Marder, T.B. Synthesis and Photophysics of a 2,7-Disubstituted Donor-Acceptor Pyrene Derivative: An Example of the Application of Sequential Ir-Catalzyed C-H Borylation and Substitution Chemistry. J. Org. Chem. 2015, 80, 5658–5665. [Google Scholar] [CrossRef] [PubMed]
- Ji, L.; Fucke, K.; Bose, S.K.; Marder, T.B. Iridium-Catalyzed Borylation of Pyrene: Irreversibility and the Influence of Ligand on Selectivity. J. Org. Chem. 2015, 80, 661–665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merz, J.; Fink, J.; Friedrich, A.; Krummenacher, I.; Al Mamari, H.H.; Lorenzen, S.; Haehnel, M.; Eichhorn, A.; Moos, M.; Holzapfel, M.; et al. Pyrene MO Shuffle—Controlling Excited State and Redox Properties by Changing the Nature of the Frontier Orbitals. Chem. Eur. J. 2017, 23, 13164–13180. [Google Scholar] [CrossRef]
- Zahid, N.I.; Ji, L.; Khyasudeen, M.F.; Friedrich, A.; Hashim, R.; Marder, T.B.; Abou-Zied, O.K. Evidence of Increased Hydrophobicity and Dynamics inside the Tail Region of Glycolipid Self-Assemblies Using 2-n-Alkyl-Pyrene Derivatives to Probe Different Locations. Langmuir 2019, 35, 9584–9592. [Google Scholar] [CrossRef]
- Merz, J.; Dietz, M.; Vonhausen, Y.; Wöber, F.; Friedrich, A.; Sieh, D.; Krummenacher, I.; Braunschweig, H.; Moos, M.; Holzapfel, M.; et al. Synthesis, Photophysical and Electronic Properties of New Red to NIR Emitting Donor-Acceptor Pyrene Derivatives. Chem. Eur. J. 2020, 26, 438–453. [Google Scholar] [CrossRef] [PubMed]
- Merz, V.; Merz, J.; Kirchner, M.; Lenhart, J.; Marder, T.B.; Krueger, A. Pyrene-Based “Turn-Off” Probe with Broad Detection Range for Cu2+, Pb2+ and Hg2+ Ions. Chem. Eur. J. 2021, 27, 8118–8126. [Google Scholar] [CrossRef]
- Wawi, M.J.; Bijoux, A.; Inguimbert, N.; Mahler, C.; Wagner, S.; Marder, T.B.; Ribou, A.C. Peptide Vectors Carry Pyrene to Cell Organelles Allowing Real-time Quantification of Free Radicals in Mitochondria by Time-resolved Fluorescence Microscopy. ChemBioChem 2021, 22, 1676–1685. [Google Scholar] [CrossRef]
- Schmitt, H.-C.; Fischer, I.; Ji, L.; Merz, J.; Marder, T.B.; Hoche, J.; Röhr, M.I.S.; Mitric, R. Isolated 2-Hydroxypyrene and its Dimer: A Frequency- and Time-Resolved Spectroscopic Study. New J. Chem. 2021, 45, 14949–14956. [Google Scholar] [CrossRef]
- Wawi, M.J.; Mahler, C.; Inguimbert, N.; Marder, T.B.; Ribou, A.-C. A new mitochondrial probe combining pyrene and a triphenylphosphonium salt for cellular oxygen and free radical detection via fluorescence lifetime measurements. Free Radic. Res. 2022, 56, 258–272. [Google Scholar] [CrossRef] [PubMed]
- Ji, L.; Riese, S.; Schmiedel, A.; Holzapfel, M.; Fest, M.; Nitsch, J.; Churchod, B.F.E.; Friedrich, A.; Wu, L.; Al Mamari, H.A.; et al. Thermodynamic Equilibrium Between Locally Excited and Charge-Transfer States through Thermally Activated Charge Transfer in 1-(Pyrene-2′-yl)-o-carborane. Chem. Sci. 2022, 13, 5205–5219. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.; Kole, G.K.; Friedrich, A.; Muller-Buschbaum, K.; Liu, Z.; Yu, X.; Marder, T.B. Comparison Study of the Site-Effect on Regioisomeric Pyridyl Pyrene Conjugates: Synthesis, Structures, and Photophysical Properties. J. Org. Chem. 2020, 85, 4256–4266. [Google Scholar] [CrossRef]
- Kole, G.K.; Merz, J.; Amar, A.; Fontaine, B.; Boucekkine, A.; Nitsch, J.; Lorenzen, S.; Friedrich, A.; Krummenacher, I.; Košćak, M.; et al. 2- and 2,7-Substituted para-N-Methylpyridinium Pyrenes: Syntheses, Molecular and Electronic Structures, Photophysical, Electrochemical, and Spectroelectrochemical Properties and Binding to Double-Stranded (ds) DNA. Eur. J. Chem. 2021, 27, 2837–2853. [Google Scholar] [CrossRef]
- Malojčić, G.; Piantanida, I.; Marinić, M.; Zinić, M.; Marjanović, M.; Kralj, M.; Pavelić, K.; Schneider, H.-J. A novel bis-phenanthridine triamine with pH controlled binding to nucleotides and nucleic acids. Org. Biomol. Chem. 2005, 3, 4373–4381. [Google Scholar] [CrossRef]
- Manallack, D.T.; Prankerd, R.J.; Yuriev, E.; Oprea, T.I.; Chalmers, D.K. The significance of acid/base properties in drug discovery. Chem. Soc. Rev. 2013, 42, 485–496. [Google Scholar] [CrossRef] [Green Version]
- Wong, P.; Lee, C.; Tannock, I.F. Reduction of intracellular pH as a strategy to enhance the pH-dependent cytotoxic effects of melphalan for human breast cancer cells. Clin. Cancer Res. 2005, 11, 3553–3557. [Google Scholar] [CrossRef] [Green Version]
- Gillies, R.J.; Robey, I.; Gatenby, R.A. Causes and consequences of increased glucose metabolism of cancers. J. Nucl. Med. 2008, 49, 24s–42s. [Google Scholar] [CrossRef] [Green Version]
- Wojtkowiak, J.W.; Verduzco, D.; Schramm, K.J.; Gillies, R.J. Drug Resistance and Cellular Adaptation to Tumor Acidic pH Microenvironment. Mol. Pharm. 2011, 8, 2032–2038. [Google Scholar] [CrossRef]
- Mergny, J.-L.; Lacroix, L. Analysis of Thermal Melting Curves. Oligonucleotides 2003, 13, 515–537. [Google Scholar] [CrossRef] [PubMed]
- Bolte, S.; Cordelieres, F.P. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 2006, 224, 213–232. [Google Scholar] [CrossRef] [PubMed]
- Piantanida, I.; Palm, B.S.; Zinic, M.; Schneider, H.J. A new 4,9-diazapyrenium intercalator for single- and double-stranded nucleic acids: Distinct differences from related diazapyrenium compounds and ethidium bromide. J. Chem. Soc. Perk. Trans. 2 2001, 9, 1808–1816. [Google Scholar] [CrossRef]
- Georghiou, S. Interaction of acridine drugs with DNA and nucleotides. Photochem. Photobiol. 1977, 26, 59–68. [Google Scholar] [CrossRef]
- Scatchard, G. The attractions of proteins for small molecules and ions. Ann. N. Y. Acad. Sci. 1949, 51, 660–672. [Google Scholar] [CrossRef]
- McGhee, J.D.; Hippel, P.H.V. Theoretical Aspects of DNA-Protein Interactions—Cooperative and Non-Cooperative Binding of Large Ligands to a One-Dimensional Homogeneous Lattice. J. Mol. Biol. 1974, 86, 469–489. [Google Scholar] [CrossRef]
- Eriksson, M.; Nordén, B. Linear and Circular Dichroism of Drug-Nucleic Acid Complexes. Meth. Enzymol. 2001, 340, 68–98. [Google Scholar]
- Šmidlehner, T.; Piantanida, I.; Pescitelli, G. Polarization spectroscopy methods in the determination of interactions of small molecules with nucleic acids—Tutorial. Beil. J. Org. Chem. 2018, 14, 84–105. [Google Scholar] [CrossRef]
- Sun, Y.; Shi, Z.; Lin, Y.; Zhang, M.; Liu, J.; Zhu, L.; Chen, Q.; Bi, J.; Li, S.; Ni, Z.; et al. Benzo(a)pyrene induces MUC5AC expression through the AhR/mitochondrial ROS/ERK pathway in airway epithelial cells. Ecotoxicol. Environ. Saf. 2021, 210, 111857. [Google Scholar] [CrossRef]
- Kang, R.-R.; Sun, Q.; Chen, K.-G.; Cao, Q.-T.; Liu, C.; Liu, K.; Ma, Z.; Deng, Y.; Liu, W.; Xu, B. Resveratrol prevents benzo(a)pyrene-induced disruption of mitochondrial homeostasis via the AMPK signaling pathway in primary cultured neurons. Environ. Pollut. 2020, 261, 114207. [Google Scholar] [CrossRef]
- Mukherjee, S.; Das Sarma, J.; Shunmugam, R. pH-Sensitive Nanoaggregates for Site-Specific Drug-Delivery as Well as Cancer Cell Imaging. ACS Omega 2016, 1, 755–764. [Google Scholar] [CrossRef] [PubMed]
- Saenger, W. Principles of Nucleic Acid Structure; Springer: New York, NY, USA, 1983. [Google Scholar]
- Cantor, C.R.; Schimmel, P.R. Biophysical Chemistry. WH Freeman Co. 1980, 3, 1109–1181. [Google Scholar]














| r | ct-DNA | Poly dAdT-Poly dAdT | pApU | |
|---|---|---|---|---|
| 1Md | 0.2 | c 2 | 8 | 3 |
| 0.3 | c 2 | 9 | 3 | |
| 2Md | 0.2 | d >16 | 31 | 4 |
| 0.3 | d >16 | 33 | - | |
| 1e | 0.1 | 0 | - | - |
| 0.3 | 2 | - | - | |
| 2e | 0.1 | 1.5 | - | - |
| ct-DNA | BSA | GC-DNA | AU-RNA | AT-DNA | ||
|---|---|---|---|---|---|---|
| 1M | pH 7 | 5.5 | 5.4 | 7.0 | 5.8 | 5.9 |
| 2M | pH 7 | 8.8 | 6.9 | 9.1 | 8.5 | 7.1 |
| 1 | pH 7 | b | 5.5 5.5 | - | - | - |
| pH 5 | 5.3 | |||||
| 2 | pH 7 | b | 5.3 5.1 | - | - | - |
| pH 5 | 5.7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Košćak, M.; Pehar, I.; Božinović, K.; Kole, G.K.; Sobočanec, S.; Podgorski, I.I.; Pinterić, M.; Müller-Buschbaum, K.; Majhen, D.; Piantanida, I.; et al. Para-N-Methylpyridinium Pyrenes: Impact of Positive Charge on ds-DNA/RNA and Protein Recognition, Photo-Induced Bioactivity, and Intracellular Localisation. Pharmaceutics 2022, 14, 2499. https://doi.org/10.3390/pharmaceutics14112499
Košćak M, Pehar I, Božinović K, Kole GK, Sobočanec S, Podgorski II, Pinterić M, Müller-Buschbaum K, Majhen D, Piantanida I, et al. Para-N-Methylpyridinium Pyrenes: Impact of Positive Charge on ds-DNA/RNA and Protein Recognition, Photo-Induced Bioactivity, and Intracellular Localisation. Pharmaceutics. 2022; 14(11):2499. https://doi.org/10.3390/pharmaceutics14112499
Chicago/Turabian StyleKošćak, Marta, Isabela Pehar, Ksenija Božinović, Goutam Kumar Kole, Sandra Sobočanec, Iva I. Podgorski, Marija Pinterić, Klaus Müller-Buschbaum, Dragomira Majhen, Ivo Piantanida, and et al. 2022. "Para-N-Methylpyridinium Pyrenes: Impact of Positive Charge on ds-DNA/RNA and Protein Recognition, Photo-Induced Bioactivity, and Intracellular Localisation" Pharmaceutics 14, no. 11: 2499. https://doi.org/10.3390/pharmaceutics14112499
APA StyleKošćak, M., Pehar, I., Božinović, K., Kole, G. K., Sobočanec, S., Podgorski, I. I., Pinterić, M., Müller-Buschbaum, K., Majhen, D., Piantanida, I., & Marder, T. B. (2022). Para-N-Methylpyridinium Pyrenes: Impact of Positive Charge on ds-DNA/RNA and Protein Recognition, Photo-Induced Bioactivity, and Intracellular Localisation. Pharmaceutics, 14(11), 2499. https://doi.org/10.3390/pharmaceutics14112499