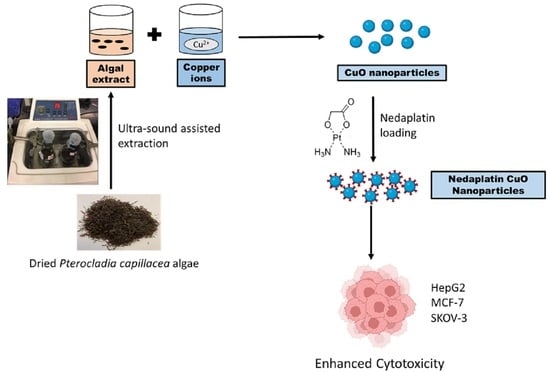
Enhanced Anticancer Activity of Nedaplatin Loaded onto Copper Nanoparticles Synthesized Using Red Algae
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Ultrasound-Assisted Extraction (UAE)
2.3. Analysis of the Algal Extracts
2.3.1. Determination of the Total Carbohydrate Content
2.3.2. Determination of the Total Phenolic Content
2.3.3. Determination of the DPPH Free Radical Scavenging Activity
2.4. Synthesis of Copper Nanoparticles
2.5. Characterization of Copper Nanoparticles
2.6. Nedaplatin Loading onto the Nanoparticles
2.6.1. In Vitro Drug Release
2.6.2. Cell Culture
2.6.3. In Vitro Cytotoxicity Tests
2.7. Statistical Analysis
3. Results and Discussion
3.1. Algal Extraction
3.2. Algal-Mediated Synthesis of CuO NPs
3.3. Nedaplatin Loading
3.4. Drug Release Study
3.5. Cytotoxicity Studies
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fahmy, S.A.; Ponte, F.; Abd El-Rahman, M.K.; Russo, N.; Sicilia, E.; Shoeib, T. Investigation of the host-guest complexation between 4-sulfocalix[4]arene and nedaplatin for potential use in drug delivery. Spectrochim. Acta—Part A Mol. Biomol. Spectrosc. 2018, 193, 528–536. [Google Scholar] [CrossRef] [PubMed]
- El-Shafie, S.; Fahmy, S.A.; Ziko, L.; Elzahed, N.; Shoeib, T.; Kakarougkas, A. Encapsulation of nedaplatin in novel pegylated liposomes increases its cytotoxicity and genotoxicity against a549 and u2os human cancer cells. Pharmaceutics 2020, 12, 863. [Google Scholar] [CrossRef] [PubMed]
- Johnstone, T.C.; Suntharalingam, K.; Lippard, S.J. The next generation of platinum drugs: Targeted pt(II) agents, nanoparticle delivery, and pt(IV) prodrugs. Chem. Rev. 2016, 116, 3436–3486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, M.; Zhang, H.; Li, H.; Li, X.; Liu, Y.; Chen, X.; Li, K. The toxicity and long-term efficacy of nedaplatin and paclitaxel treatment as neoadjuvant chemotherapy for locally advanced cervical cancer. J. Surg. Oncol. 2012, 105, 206–211. [Google Scholar] [CrossRef]
- Xu, J.; He, X.; Cheng, K.; Guo, W.; Bian, X.; Jiang, X.; Huang, S. Concurrent chemotherapy with nedaplatin plus paclitaxel or fluorouracil for locoregionally advanced nasopharyngeal carcinoma: Survival and toxicity. Head Neck 2014, 36, 1474–1480. [Google Scholar] [CrossRef]
- Fahmy, S.A.; Fawzy, I.M.; Saleh, B.M.; Issa, M.Y.; Bakowsky, U.; Azzazy, H.M.E. Green Synthesis of Platinum and Palladium Nanoparticles Using Peganum harmala L. Seed Alkaloids: Biological and Computational Studies. Nanomaterials 2021, 11, 965. [Google Scholar] [CrossRef]
- Fahmy, S.A.; Issa, M.Y.; Saleh, B.M.; Meselhy, M.R.; Azzazy, H.M.E. Peganum harmala alkaloids self-assembled supramolecular nanocapsules with enhanced antioxidant and cytotoxic activities. ACS Omega 2021, 6, 11954–11963. [Google Scholar] [CrossRef]
- Fahmy, S.A.; Brüßler, J.; Ponte, F.; Abd El-Rahman, M.K.; Russo, N.; Sicilia, E.; Bakowsky, U.; Shoeib, T. A study on the physicochemical properties and cytotoxic activity of p-sulfocalix[4]arene-nedaplatin complex. J. Phys. Conf. Ser. 2019, 1310, 012011. [Google Scholar] [CrossRef]
- Fahmy, S.A.; Brüßler, J.; Alawak, M.; El-Sayed, M.M.H.; Bakowsky, U.; Shoeib, T. Chemotherapy based on supramolecular chemistry: A promising strategy in cancer therapy. Pharmaceutics 2019, 11, 292. [Google Scholar] [CrossRef] [Green Version]
- Fahmy, S.A.; Ramzy, A.; Saleh, B.M.; Azzazy, H.M.E. Stimuli-Responsive Amphiphilic Pillar[n]arene Nanovesicles for Targeted Delivery of Cancer Drugs. ACS Omega 2021, 6, 25876–25883. [Google Scholar] [CrossRef]
- Fahmy, S.A.; Azzazy, H.M.E.; Schaefer, J. Liposome Photosensitizer Formulations for Effective Cancer Photodynamic Therapy. Pharmaceutics 2021, 13, 1345. [Google Scholar] [CrossRef] [PubMed]
- Fahmy, S.A.; Preis, E.; Bakowsky, U.; Azzazy, H.M.E.-S. Palladium Nanoparticles Fabricated by Green Chemistry: Promising Chemotherapeutic, Antioxidant and Antimicrobial Agents. Materials 2020, 13, 3661. [Google Scholar] [CrossRef] [PubMed]
- Fahmy, S.A.; Preis, E.; Bakowsky, U.; Azzazy, H.M.E. Platinum Nanoparticles: Green Synthesis and Biomedical Applications. Molecules 2020, 25, 4981. [Google Scholar] [CrossRef]
- Gu, H.; Chen, X.; Chen, F.; Zhou, X.; Parsaee, Z. Ultrasound-assisted biosynthesis of CuO-NPs using brown alga cystoseira trinodis: Characterization, photocatalytic AOP, DPPH scavenging and antibacterial investigations. Ultrason.—Sonochem. 2018, 41, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Netanel Liberman, G.; Ochbaum, G.; Arad, S.; Bitton, R. The sulfated polysaccharide from a marine red microalga as a platform for the incorporation of zinc ions. Carbohydr. Polym. 2016, 152, 658–664. [Google Scholar] [CrossRef]
- Patil, M.P.; Kim, G. Marine microorganisms for synthesis of metallic nanoparticles and their biomedical applications. Colloids Surf. B Biointerfaces 2018, 172, 487–495. [Google Scholar] [CrossRef]
- Essa, H.; Fleita, D.; Rifaat, D.; Samy, S.; El-Sayed, M.M.H. Towards optimizing the conventional and ultrasonic-assisted extraction of sulphated polysaccharides from marine algae. IOP Conf. Ser. Mater. Sci. Eng. 2018, 464, 012007. [Google Scholar] [CrossRef]
- El Azm, N.A.; Fleita, D.; Rifaat, D.; Mpingirika, E.Z.; Amleh, A.; El-Sayed, M.M.H. Production of bioactive compounds from the sulfated polysaccharides extracts of Ulva lactuca: Post-extraction enzymatic hydrolysis followed by ion-exchange chromatographic fractionation. Molecules 2019, 24, 2132. [Google Scholar] [CrossRef] [Green Version]
- Essa, H.L.; Abdelfattah, M.S.; Marzouk, A.S.; Shedeed, Z.; Guirguis, H.A.; El-Sayed, M.M.H. Biogenic copper nanoparticles from Avicennia marina leaves: Impact on seed germination, detoxification enzymes, chlorophyll content and uptake by wheat seedlings. PLoS ONE 2021, 16, e0249764. [Google Scholar] [CrossRef]
- Fahmy, S.A.; Mamdouh, W. Garlic oil–loaded PLGA nanoparticles with controllable size and shape and enhanced antibacterial activities. J. Appl. Polym. Sci. 2018, 135, 46133. [Google Scholar] [CrossRef]
- Azzazy, H.M.E.; Fahmy, S.A.; Mahdy, N.K.; Meselhy, M.R.; Bakowsky, U. Chitosan-Coated PLGA Nanoparticles Loaded with Peganum harmala Alkaloids with Promising Antibacterial and Wound Healing Activities. Nanomaterials 2021, 11, 2438. [Google Scholar] [CrossRef] [PubMed]
- Fahmy, S.A.; Ponte, F.; Fawzy, I.M.; Sicilia, E.; Bakowsky, U.; Azzazy, H.M.E. Betaine host–guest complexation with a calixarene receptor: Enhanced in vitro anticancer effect. RSC Adv. 2021, 11, 24673–24680. [Google Scholar] [CrossRef]
- Fahmy, S.A.; Ponte, F.; Sicilia, E.; Azzazy, H.M.E. Experimental and Computational Investigations of Carboplatin Supramolecular Complexes. ACS Omega 2020, 5, 31456–31466. [Google Scholar] [CrossRef] [PubMed]
- Fahmy, S.A.; Ponte, F.; Fawzy, I.M.; Sicilia, E.; Bakowsky, U.; Azzazy, H.M.E. Host-Guest Complexation of Oxaliplatin and Para-Sulfonatocalix[n]Arenes for Potential Use in Cancer Therapy. Molecules 2020, 25, 5926. [Google Scholar] [CrossRef]
- Obluchinsksya, E.D.; Makarova, M.N.; Pozharitskaya, O.N.; Shikov, A.N. Effects of ultrasound treatment on the chemical composition and anticoagulant properties of dry Fucus extract. Pharm. Chem. J. 2015, 49, 183–186. [Google Scholar] [CrossRef]
- Hmelkov, A.B.; Zvyagintseva, T.N.; Zvyagintseva, T.N.; Shevchenko, N.M.; Shevchenko, N.M.; Ermakova, S.P. Ultrasound-assisted extraction of polysaccharides from brown alga fucus evanescens. structure and biological activity of the new fucoidan fractions. J. Appl. Phycol. 2018, 30, 2039–2046. [Google Scholar] [CrossRef]
- Rahimi, F.; Tabarsa, M.; Rezaei, M. Ulvan from green algae ulva intestinalis: Optimization of ultrasound-assisted extraction and antioxidant activity. J. Appl. Phycol. 2016, 28, 2979–2990. [Google Scholar] [CrossRef]
- Dang, T.T.; Van Vuong, Q.; Schreider, M.J.; Bowyer, M.C.; Van Altena, I.A.; Scarlett, C.J. Optimisation of ultrasound-assisted extraction conditions for phenolic content and antioxidant activities of the alga hormosira banksii using response surface methodology. J. Appl. Phycol. 2017, 29, 3161–3173. [Google Scholar] [CrossRef]
- Essa, H.L.; Guirguis, H.A.; El-Sayed, M.M.H.; Rifaat, D.; Abdelfattah, M.S. Ultrasonically-extracted marine polysaccharides as potential green antioxidant alternatives. Proceedings 2020, 67, 23. [Google Scholar] [CrossRef]
- Zhao, G.; Chen, X.; Wang, L.; Zhou, S.; Feng, H.; Chen, W.N.; Lau, R. Ultrasound assisted extraction of carbohydrates from microalgae as feedstock for yeast fermentation. Bioresour. Technol. 2013, 128, 337–344. [Google Scholar] [CrossRef]
- El-Sayed, M.M.H.; Fleita, D.; Rifaat, D.; Essa, H. Assessment of the state-of-the-art developments in the extraction of antioxidants from marine algal species. In Ingredients Extraction by Physicochemical Methods in Food; Grumezescu, A.M., Holban, A.M., Eds.; Academic Press: Cambridge, MA, USA; Elsevier: Cambridge, MA, USA, 2018; Chapter 9; pp. 367–397. [Google Scholar]
- Fleita, D.; El-Sayed, M.; Rifaat, D. Evaluation of the antioxidant activity of enzymatically-hydrolyzed sulfated polysaccharides extracted from red algae; Pterocladia capillacea. LWT—Food Sci. Technol. 2015, 63, 1236–1244. [Google Scholar] [CrossRef]
- Garcia-Vaquero, M.; Rajauria, G.; Tiwari, B.; Sweeney, T.; O’Doherty, J. Extraction and Yield Optimisation of Fucose, Glucans and Associated Antioxidant Activities from Laminaria digitata by Applying Response Surface Methodology to High Intensity Ultrasound-Assisted Extraction. Mar. Drugs 2018, 16, 257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arya, A.; Gupta, K.; Chundawat, T.S.; Vaya, D. Biogenic synthesis of copper and silver nanoparticles using green alga botryococcus braunii and its antimicrobial activity. Bioinorg. Chem. Appl. 2018, 3, 7879403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaikh, R.R.; Mirza, S.S.; Sawant, M.R.; Dare, S.B. Biosynthesis of Copper Nanoparticles using Vitis vinifera Leaf Extract and Its Antimicrobial Activity. Pharm. Lett. 2016, 8, 265–272. [Google Scholar]
- Khandel, P.; Yadaw, R.K.; Soni, D.K.; Kanwar, L.; Shahi, S.K. Biogenesis of metal nanoparticles and their pharmacological applications: Present status and application prospects. J. Nanostruct. Chem. 2018, 8, 217–254. [Google Scholar] [CrossRef] [Green Version]
- Sarkar, J.; Chakraborty, N.; Chatterjee, A.; Bhattacharjee, A.; Dasgupta, D.; Acharya, K. Green Synthesized Copper Oxide Nanoparticles Ameliorate Defence and Antioxidant Enzymes in Lens culinaris. Nanomaterials 2020, 10, 312. [Google Scholar] [CrossRef] [Green Version]
- Ismail, M. Green synthesis and characterizations of copper nanoparticles. Mater. Chem. Phys. 2020, 240, 122283. [Google Scholar] [CrossRef]
- Raveendran, S.; Poulose, A.C.; Yoshida, Y.; Maekawa, T.; Kumar, D.S. Bacterial exopolysaccharide based nanoparticles for sustained drug delivery, cancer chemotherapy and bioimaging. Carbohydr. Polym. 2013, 91, 22–32. [Google Scholar] [CrossRef]
- Posocco, B.; Dreussi, E.; De Santa, J.; Toffoli, G.; Abrami, M.; Musiani, F.; Grassi, M.; Farra, R.; Tonon, F.; Grassi, G.; et al. Polysaccharides for the Delivery of Antitumor Drugs. Materials 2015, 8, 2569–2615. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.W.; Jeong, D.; Na, K. Doxorubicin loading fucoidan acetate nanoparticles for immune and chemotherapy in cancer treatment. Carbohydr. Polym. 2013, 94, 850–856. [Google Scholar] [CrossRef]
- Chollet, L.; Saboural, P.; Chauvierre, C.; Villemin, J.-N.; Letourneur, D.; Chaubet, F. Fucoidans in Nanomedicine. Mar. Drugs 2016, 14, 145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cunha, L.; Grenha, A. Sulfated seaweed polysaccharides as multifunctional materials in drug delivery applications. Mar. Drugs 2016, 14, 42. [Google Scholar] [CrossRef]
- Erfani, N.; Nazemosadat, Z.; Moein, M. Cytotoxic activity of ten algae from the Persian Gulf and Oman Sea on human breast cancer cell lines; MDA-MB-231, MCF-7, and T-47D. Pharmacogn. Res. 2015, 7, 133–137. [Google Scholar] [CrossRef] [Green Version]
- Mashjoor, S.; Yousefzadi, M.; Esmaeili, M.A.; Rafiee, R. Cytotoxicity and antimicrobial activity of marine macro algae (Dictyotaceae and Ulvaceae) from the Persian Gulf. Cytotechnology 2016, 68, 1717–1726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mansur, A.A.; Brown, M.T.; Billington, R.A. The cytotoxic activity of extracts of the brown alga Cystoseira tamariscifolia (Hudson) Papenfuss, against cancer cell lines changes seasonally. J. Appl. Phycol. 2020, 32, 2419–2429. [Google Scholar] [CrossRef]
- Kesavan, A.; Ilaiyaraja, P.; Beaula, W.S.; Kumari, V.V.; Lal, J.S.; Arunkumar, C.; Venkatraman, G. Tumor targeting using polyamidoamine dendrimer–cisplatin nanoparticles functionalized with diglycolamic acid and herceptin. Eur. J. Pharm. Biopharm. 2015, 96, 255–263. [Google Scholar] [CrossRef] [PubMed]
- Aldawsari, H.M.; Singh, S. Rapid Microwave-Assisted Cisplatin-Loaded Solid Lipid Nanoparticles: Synthesis, Characterization and Anticancer Study. Nanomaterials 2020, 10, 510. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Meng, Z.; Li, Q.; Huang, X.; Kang, Z.; Dong, H.; Li, C. A pH responsive complexation-based drug delivery system for oxaliplatin. Chem. Sci. 2017, 8, 4458–4464. [Google Scholar] [CrossRef] [Green Version]
- Jia, C.; Zhong, Y.; Zhang, X.; Liao, X.; Li, Y.; Yang, B.; Gao, C. Host–guest inclusion systems of nedaplatin with cucurbit[7]uril for improved in vitro antitumour activity. J. Incl. Phenom. Macrocycl. Chem. 2020, 97, 99–107. [Google Scholar] [CrossRef]




| Time, h | % Yield | % Carbohydrate | TPC, mgGA/g |
|---|---|---|---|
| 0.5 | 14.2 ± 0.1 | 24.08 ± 0.03 | 12.36 ± 0.03 |
| 1 | 15.10 ± 0.03 | 46.6 ± 0.4 | 9.72 ± 0.03 |
| 2 | 16.10 ± 0.04 | 59.3 ± 0.2 | 9.48 ± 0.04 |
| 4 | 16.08 ± 0.05 | 55.1 ± 0.1 | 9.44 ± 0.06 |
| Functional Group | Wavenumber (cm−1) | Bond | Extract | CuO NPs |
|---|---|---|---|---|
| Hydroxyl group | 3500–3000 | O-H | √ | √ |
| Amide group | 1670–1600 | C=O | √ | √ |
| Sulfate | 1450–1350 | S=O | √ | √ |
| Acidic polysaccharide | 1120–1000 | √ | √ | |
| Ester sulfate | 805–900 | C-O-S | √ | √ |
| Aromatic ester | 1310–1250 | C=O | √ | X |
| Platinum-Based Drug | Carrier | Cancer Cells | IC50 (µg/mL) | Reference |
|---|---|---|---|---|
| Cisplatin | Herceptin targeted, diglycolamic acid (DGA) functionalized polyamidoamine (PAMAM) dendrimer | SKOV-3 | 6.6 | [47] |
| Cisplatin | Solid lipid nanoparticle | MCF-7 | 6.51 | [48] |
| Oxaliplatin | Carboxylato-pillar [6] arene | HEP-G2 | 7.6 | [49] |
| Nedaplatin | Cucurbit [7] uril | MCF-7 | 11.8 | [50] |
| Nedaplatin | CuO NPs | HEP-G2 MCF-7 SKOV-3 | 0.4 1.5 0.7 | This study |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aboeita, N.M.; Fahmy, S.A.; El-Sayed, M.M.H.; Azzazy, H.M.E.-S.; Shoeib, T. Enhanced Anticancer Activity of Nedaplatin Loaded onto Copper Nanoparticles Synthesized Using Red Algae. Pharmaceutics 2022, 14, 418. https://doi.org/10.3390/pharmaceutics14020418
Aboeita NM, Fahmy SA, El-Sayed MMH, Azzazy HME-S, Shoeib T. Enhanced Anticancer Activity of Nedaplatin Loaded onto Copper Nanoparticles Synthesized Using Red Algae. Pharmaceutics. 2022; 14(2):418. https://doi.org/10.3390/pharmaceutics14020418
Chicago/Turabian StyleAboeita, Nada Mostafa, Sherif Ashraf Fahmy, Mayyada M. H. El-Sayed, Hassan Mohamed El-Said Azzazy, and Tamer Shoeib. 2022. "Enhanced Anticancer Activity of Nedaplatin Loaded onto Copper Nanoparticles Synthesized Using Red Algae" Pharmaceutics 14, no. 2: 418. https://doi.org/10.3390/pharmaceutics14020418
APA StyleAboeita, N. M., Fahmy, S. A., El-Sayed, M. M. H., Azzazy, H. M. E. -S., & Shoeib, T. (2022). Enhanced Anticancer Activity of Nedaplatin Loaded onto Copper Nanoparticles Synthesized Using Red Algae. Pharmaceutics, 14(2), 418. https://doi.org/10.3390/pharmaceutics14020418