Squalene-Based Nano-Assemblies Improve the Pro-Autophagic Activity of Trehalose
Abstract
:1. Introduction
2. Results
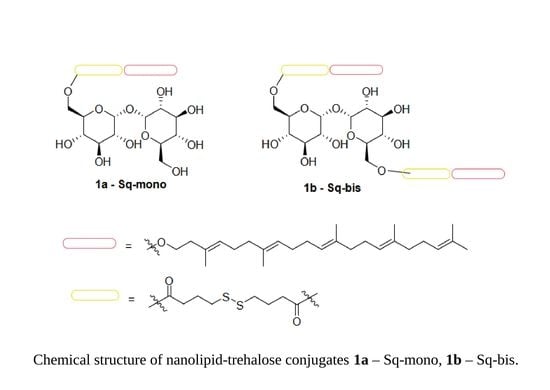
2.1. Synthesis of Mono- and Bis-Squalene-Trehalose Conjugates
2.2. Assembly of Mono- and Bis-Squalene-Trehalose NAs
2.3. Biological Characterization of Trehalose-Based Small Molecules and NAs: Toxicity
2.4. Biological Characterization of Trehalose-Based Small Molecules and NAs: Efficacy
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Synthesis
5.1.1. General
5.1.2. Synthesis of Mono Dithio-Dibutyroate Squalenoyl-hexaTMS-Trehalose 4a and Bis Dithio-Dibutyroate Squalenoyl-hexaTMS-Trehalose 4b
5.1.3. Synthesis of Mono Dithio-Dibutyroate Squalenoyl-Trehalose 1a
5.1.4. Synthesis of Bis Dithio-Dibutyroate Squalenoyl-Trehalose 1b
5.2. Cell Cultures
5.3. Cytotoxicity Assay
5.4. Autophagy Assay
5.5. Autophagosome–Lysosome Fusion Assay
5.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Deleidi, M.; Maetzler, W. Protein clearance mechanisms of alpha-synuclein and amyloid-Beta in lewy body disorders. Int. J. Alzheimers. Dis. 2012, 2012, 391438. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.; Ambasta, R.K.; Kumar, P. Ubiquitin biology in neurodegenerative disorders: From impairment to therapeutic strategies. Ageing Res. Rev. 2020, 61, 101078. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Wang, J.; Yang, C. Celastrol, a TFEB (transcription factor EB) agonist, is a promising drug candidate for Alzheimer disease. Autophagy 2022, 6, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Pischedda, F.; Cirnaru, M.D.; Ponzoni, L.; Sandre, M.; Biosa, A.; Carrion, M.P.; Marin, O.; Morari, M.; Pan, L.; Greggio, E.; et al. LRRK2 G2019S kinase activity triggers neurotoxic NSF aggregation. Brain 2021, 144, 1509–1525. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.A.; Rhim, H. Therapeutic Implication of Autophagy in Neurodegenerative Diseases. BMB Rep. 2017, 50, 345–354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rahman, M.A.; Cho, Y.; Nam, G.; Rhim, H. Antioxidant Compound, Oxyresveratrol, Inhibits APP Production through the AMPK/ULK1/mTOR mediated Autophagy Pathway in Mouse Cortical Astrocytes. Antioxidants 2021, 10, 108. [Google Scholar] [CrossRef] [PubMed]
- White, E. The Role for Autophagy in Cancer. J. Clin. Investig. 2015, 125, 42–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mrakovcic, M.; Fröhlich, L.F. p53-Mediated Molecular Control of Autophagy in Tumor Cells. Biomolecules 2018, 8, 14. [Google Scholar] [CrossRef] [Green Version]
- Khan, A.; Singh, V.K.; Thakral, D.; Gupta, R. Autophagy in acute myeloid leukemia: A paradoxical role in chemoresistance. Clin. Transl. Oncol. 2022; ahead of print. [Google Scholar] [CrossRef]
- Klionsky, D.J. Autophagy in major human diseases. EMBO J. 2021, 40, e108863. [Google Scholar] [CrossRef]
- Mulcahy Levy, J.M.; Thorburn, A. Autophagy in cancer: Moving from understanding mechanism to improving therapy responses in patients. Cell Death Differ. 2020, 27, 843–857. [Google Scholar] [CrossRef]
- Khan, A.A.; Stocker, B.L.; Timmer, M.S.M. Trehalose glycolipids--synthesis and biological activities. Carbohydr. Res. 2012, 356, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Crowe, J.H.; Crowe, L.M.; Chapman, D. Preservation of membranes in anhydrobiotic organisms: The role of trehalose. Science 1984, 223, 701–703. [Google Scholar] [CrossRef] [PubMed]
- Argüelles, J.-C. Why can’t vertebrates synthesize trehalose? J. Mol. Evol. 2014, 79, 111–116. [Google Scholar] [CrossRef] [PubMed]
- Abazari, A.; Meimetis, L.G.; Budin, G.; Bale, S.S.; Weissleder, R.; Toner, M. Engineered Trehalose Permeable to Mammalian Cells. PLoS ONE 2015, 10, e0130323. [Google Scholar] [CrossRef] [PubMed]
- Stacchiotti, A.; Corsetti, G. Natural Compounds and Autophagy: Allies against Neurodegeneration. Front. Cell Dev. Biol. 2020, 8, 555409. [Google Scholar] [CrossRef]
- DeBosch, B.J.; Heitmeier, M.R.; Mayer, A.L.; Higgins, C.B.; Crowley, J.R.; Kraft, T.E.; Chi, M.; Newberry, E.P.; Chen, Z.; Finck, B.N.; et al. Trehalose inhibits solute carrier 2A (SLC2A) proteins to induce autophagy and prevent hepatic steatosis. Sci. Signal 2016, 9, ra21. [Google Scholar] [CrossRef] [Green Version]
- Rusmini, P.; Cortese, K.; Crippa, V.; Cristofani, R.; Cicardi, M.E.; Ferrari, V.; Vezzoli, G.; Tedesco, B.; Meroni, M.; Messi, E.; et al. Trehalose induces autophagy via lysosomal-mediated TFEB activation in models of motoneuron degeneration. Autophagy 2019, 15, 631–651. [Google Scholar] [CrossRef]
- Assoni, G.; Frapporti, G.; Colombo, E.; Gornati, D.; Perez-Carrion, M.D.; Polito, L.; Seneci, P.; Piccoli, G.; Arosio, D. Trehalose-based neuroprotective autophagy inducers. Bioorg. Med. Chem. Lett. 2021, 40, 127929. [Google Scholar] [CrossRef]
- Borrelli, S.; Christodoulou, M.S.; Ficarra, I.; Silvani, A.; Cappelletti, G.; Cartelli, D.; Damia, G.; Ricci, F.; Zucchetti, M.; Dosio, F.; et al. New class of squalene-based releasable nanoassemblies of paclitaxel, podophyllotoxin, camptothecin and epothilone A. Eur. J. Med. Chem. 2014, 85, 179–190. [Google Scholar] [CrossRef] [Green Version]
- Colombo, E.; Biocotino, M.; Frapporti, G.; Randazzo, P.; Christodoulou, M.S.; Piccoli, G.; Polito, L.; Seneci, P.; Passarella, D. Nanolipid-trehalose conjugates and nano-assemblies as putative autophagy inducers. Pharmaceutics 2019, 11, E422. [Google Scholar] [CrossRef] [Green Version]
- Yoshimori, T.; Yamamoto, A.; Moriyama, Y.; Futai, M.; Tashiro, Y. Bafilomycin A1, a specific inhibitor of vacuolar-type H+-ATPase, inhibits acidification and protein degradation in lysosomes of cultured cells. J. Biol. Chem. 1991, 266, 17707–17712. [Google Scholar] [CrossRef]
- Kimura, S.; Noda, T.; Yoshimori, T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 2007, 3, 452–460. [Google Scholar] [CrossRef] [Green Version]
- Pankiv, S.; Clausen, T.H.; Lamark, T.; Brech, A.; Bruun, J.-A.; Outzen, H.; Øvervatn, A.; Bjørkøy, G.; Johansen, T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 2007, 282, 24131–24145. [Google Scholar] [CrossRef] [Green Version]
- Griffith, O.W. Biologic and pharmacologic regulation of mammalian glutathione synthesis. Free Radic. Biol. Med. 1999, 27, 922–935. [Google Scholar] [CrossRef]
- Kalf, G.F.; Rieder, S.V. The purification and properties of trehalase. J. Biol. Chem. 1958, 230, 691–698. [Google Scholar] [CrossRef]
- Adhikari, P.; Pal, P.; Das, A.K.; Ray, S.; Bhattacharjee, A.; Mazumder, B. Nano lipid-drug conjugate: An integrated review. Int. J. Pharm. 2017, 529, 629–641. [Google Scholar] [CrossRef]
- Couvreur, P.; Stella, B.; Reddy, L.H.; Hillaireau, H.; Dubernet, C.; Desmaële, D.; Lepêtre-Mouelhi, S.; Rocco, F.; Dereuddre-Bosquet, N.; Clayette, P.; et al. Squalenoyl nanomedicines as potential therapeutics. Nano Lett. 2006, 6, 2544–2548. [Google Scholar] [CrossRef]
- Sémiramoth, N.; Di Meo, C.; Zouhiri, F.; Saïd-Hassane, F.; Valetti, S.; Gorges, R.; Nicolas, V.; Poupaert, J.H.; Chollet-Martin, S.; Desmaële, D.; et al. Self-assembled squalenoylated penicillin bioconjugates: An original approach for the treatment of intracellular infections. ACS Nano 2012, 6, 3820–3831. [Google Scholar] [CrossRef]
- Sui, X.; Chen, R.; Wang, Z.; Huang, Z.; Kong, N.; Zhang, M.; Han, W.; Lou, F.; Yang, J.; Zhang, Q.; et al. Autophagy and chemotherapy resistance: A promising therapeutic target for cancer treatment. Cell Death Dis. 2013, 4, e838. [Google Scholar] [CrossRef]
- Chavez-Dominguez, R.; Perez-Medina, M.; Lopez-Gonzalez, J.S.; Galicia-Velasco, M.; Aguilar-Cazares, D. The Double-Edge Sword of Autophagy in Cancer: From Tumor Suppression to Pro-tumor Activity. Front. Oncol. 2020, 10, 578418. [Google Scholar] [CrossRef]
- Battaglia, L.; Gallarate, M. Lipid nanoparticles: State of the art, new preparation methods and challenges in drug delivery. Expert Opin. Drug Deliv. 2012, 9, 497–508. [Google Scholar] [CrossRef]









| Compound (NA) (mg/mL) | Polydispersity Index (PI) | Hydrodynamic Diameter (HD, nm) | Z-Potential (mV) |
|---|---|---|---|
| 5a (0.6 mg/mL) | 0.167 ± 0.018 | 188 ± 49 | −37.13 ± 0.85 |
| 5b (0.6 mg/mL) | 0.074 ± 0.013 | 184 ± 36 | −43.55 ± 1.60 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Frapporti, G.; Colombo, E.; Ahmed, H.; Assoni, G.; Polito, L.; Randazzo, P.; Arosio, D.; Seneci, P.; Piccoli, G. Squalene-Based Nano-Assemblies Improve the Pro-Autophagic Activity of Trehalose. Pharmaceutics 2022, 14, 862. https://doi.org/10.3390/pharmaceutics14040862
Frapporti G, Colombo E, Ahmed H, Assoni G, Polito L, Randazzo P, Arosio D, Seneci P, Piccoli G. Squalene-Based Nano-Assemblies Improve the Pro-Autophagic Activity of Trehalose. Pharmaceutics. 2022; 14(4):862. https://doi.org/10.3390/pharmaceutics14040862
Chicago/Turabian StyleFrapporti, Giulia, Eleonora Colombo, Hazem Ahmed, Giulia Assoni, Laura Polito, Pietro Randazzo, Daniela Arosio, Pierfausto Seneci, and Giovanni Piccoli. 2022. "Squalene-Based Nano-Assemblies Improve the Pro-Autophagic Activity of Trehalose" Pharmaceutics 14, no. 4: 862. https://doi.org/10.3390/pharmaceutics14040862
APA StyleFrapporti, G., Colombo, E., Ahmed, H., Assoni, G., Polito, L., Randazzo, P., Arosio, D., Seneci, P., & Piccoli, G. (2022). Squalene-Based Nano-Assemblies Improve the Pro-Autophagic Activity of Trehalose. Pharmaceutics, 14(4), 862. https://doi.org/10.3390/pharmaceutics14040862