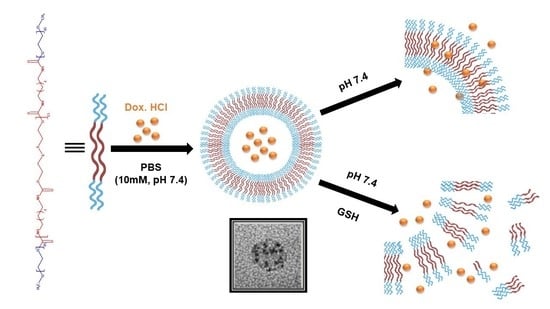
Redox-Responsive Polymersomes as Smart Doxorubicin Delivery Systems
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
2.2.1. Preparation of Blank Polymersomes
2.2.2. Morphology, Particle Size and Zeta Potential Measurements
2.2.3. Preparation and Characterization of Drug-Loaded Polymersomes
2.2.4. Calculation of Drug Encapsulation Efficiency and Drug Loading Content
2.2.5. In Vitro Drug Release Study
2.2.6. Statistical Analysis
3. Results and Discussion
3.1. Preparation and Characterization of Blank Polymersomes
3.2. Preparation and Characterization of Drug-Loaded Polymersomes
3.3. Calculation of Drug Encapsulation Efficiency and Drug Loading Content
3.4. In Vitro Drug Release Study
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Albuquerque, L.J.C.; Sincari, V.; Jäger, A.; Kucka, J.; Humajova, J.; Pankrac, J.; Paral, P.; Heizer, T.; Janouškova, O.; Davidovich, I.; et al. pH-Responsive Polymersome-Mediated Delivery of Doxorubicin into Tumor Sites Enhances the Therapeutic Efficacy and Reduces Cardiotoxic Effects. J. Control. Release 2021, 332, 529–538. [Google Scholar] [CrossRef] [PubMed]
- Sanson, C.; Schatz, C.; Le Meins, J.F.; Soum, A.; Thévenot, J.; Garanger, E.; Lecommandoux, S. A Simple Method to Achieve High Doxorubicin Loading in Biodegradable Polymersomes. J. Control. Release 2010, 147, 428–435. [Google Scholar] [CrossRef] [PubMed]
- Yassin, M.A.; Appelhans, D.; Wiedemuth, R.; Formanek, P.; Boye, S.; Lederer, A.; Temme, A.; Voit, B. Overcoming Concealment Effects of Targeting Moieties in the PEG Corona: Controlled Permeable Polymersomes Decorated with Folate-Antennae for Selective Targeting of Tumor Cells. Small 2015, 11, 1580–1591. [Google Scholar] [CrossRef]
- Zhao, L.; Zhang, X.; Liu, X.; Li, J.; Luan, Y. pH-Responsive Poly(Ethylene Glycol)-Poly(ϵ-Caprolactone)-Poly(Glutamic Acid) Polymersome as an Efficient Doxorubicin Carrier for Cancer Therapy. Polym. Int. 2017, 66, 1579–1586. [Google Scholar] [CrossRef]
- Zhu, D.; Wu, S.; Hu, C.; Chen, Z.; Wang, H.; Fan, F.; Qin, Y.; Wang, C.; Sun, H.; Leng, X.; et al. Folate-Targeted Polymersomes Loaded with Both Paclitaxel and Doxorubicin for the Combination Chemotherapy of Hepatocellular Carcinoma. Acta Biomater. 2017, 58, 399–412. [Google Scholar] [CrossRef] [PubMed]
- Aibani, N.; Nesbitt, H.; Marino, N.; Jurek, J.; O’Neill, C.; Martin, C.; Di Bari, I.; Sheng, Y.; Logan, K.; Hawthorne, S.; et al. Electroneutral Polymersomes for Combined Cancer Chemotherapy. Acta Biomater. 2018, 80, 327–340. [Google Scholar] [CrossRef]
- Alibolandi, M.; Abnous, K.; Mohammadi, M.; Hadizadeh, F.; Sadeghi, F.; Taghavi, S.; Jaafari, M.R.; Ramezani, M. Extensive Preclinical Investigation of Polymersomal Formulation of Doxorubicin versus Doxil-Mimic Formulation. J. Control. Release 2017, 264, 228–236. [Google Scholar] [CrossRef]
- Araste, F.; Aliabadi, A.; Abnous, K.; Taghdisi, S.M.; Ramezani, M.; Alibolandi, M. Self-Assembled Polymeric Vesicles: Focus on Polymersomes in Cancer Treatment. J. Control. Release 2021, 330, 502–528. [Google Scholar] [CrossRef]
- Xu, J.; Zhu, X.; Qiu, L. Polyphosphazene Vesicles for Co-Delivery of Doxorubicin and Chloroquine with Enhanced Anticancer Efficacy by Drug Resistance Reversal. Int. J. Pharm. 2016, 498, 70–81. [Google Scholar] [CrossRef]
- Li, J.; Li, Y.; Wang, Y.; Ke, W.; Chen, W.; Wang, W.; Ge, Z. Polymer Prodrug-Based Nanoreactors Activated by Tumor Acidity for Orchestrated Oxidation/Chemotherapy. Nano Lett. 2017, 17, 6983–6990. [Google Scholar] [CrossRef]
- Aibani, N.; Khan, T.N.; Callan, B. Liposome Mimicking Polymersomes; A Comparative Study of the Merits of Polymersomes in Terms of Formulation and Stability. Int. J. Pharm. X 2020, 2, 100040. [Google Scholar] [CrossRef] [PubMed]
- Alibolandi, M.; Ramezani, M.; Abnous, K.; Sadeghi, F.; Atyabi, F.; Asouri, M.; Ahmadi, A.A.; Hadizadeh, F. In Vitro and in Vivo Evaluation of Therapy Targeting Epithelial-Cell Adhesion-Molecule Aptamers for Non-Small Cell Lung Cancer. J. Control. Release 2015, 209, 88–100. [Google Scholar] [CrossRef] [PubMed]
- Hasannia, M.; Aliabadi, A.; Abnous, K.; Taghdisi, S.M.; Ramezani, M.; Alibolandi, M. Synthesis of Block Copolymers Used in Polymersome Fabrication: Application in Drug Delivery. J. Control. Release 2022, 341, 95–117. [Google Scholar] [CrossRef] [PubMed]
- Nehate, C.; Nayal, A.; Koul, V. Redox Responsive Polymersomes for Enhanced Doxorubicin Delivery. ACS Biomater. Sci. Eng. 2019, 5, 70–80. [Google Scholar] [CrossRef]
- Haas, S.; Hain, N.; Raoufi, M.; Handschuh-Wang, S.; Wang, T.; Jiang, X.; Schönherr, H. Enzyme Degradable Polymersomes from Hyaluronic Acid-Block-Poly(ε-Caprolactone) Copolymers for the Detection of Enzymes of Pathogenic Bacteria. Biomacromolecules 2015, 16, 832–841. [Google Scholar] [CrossRef]
- Lale, S.V.; Kumar, A.; Prasad, S.; Bharti, A.C.; Koul, V. Folic Acid and Trastuzumab Functionalized Redox Responsive Polymersomes for Intracellular Doxorubicin Delivery in Breast Cancer. Biomacromolecules 2015, 16, 1736–1752. [Google Scholar] [CrossRef]
- Liu, F.; Kozlovskaya, V.; Medipelli, S.; Xue, B.; Ahmad, F.; Saeed, M.; Cropek, D.; Kharlampieva, E. Temperature-Sensitive Polymersomes for Controlled Delivery of Anticancer Drugs. Chem. Mater. 2015, 27, 7945–7956. [Google Scholar] [CrossRef]
- Saravanakumar, G.; Park, H.; Kim, J.; Park, D.; Lim, J.; Lee, J.; Kim, W.J. Polymersomes with Singlet Oxygen-Labile Poly(β-Aminoacrylate) Membrane for NIR Light-Controlled Combined Chemo-Phototherapy. J. Control. Release 2020, 327, 627–640. [Google Scholar] [CrossRef]
- Tsai, M.F.; Lo, Y.L.; Huang, Y.C.; Yu, C.C.; Wu, Y.T.; Su, C.H.; Wang, L.F. Multi-Stimuli-Responsive Dox Released from Magnetosome for Tumor Synergistic Theranostics. Int. J. Nanomed. 2020, 15, 8623–8639. [Google Scholar] [CrossRef]
- Wei, P.; Sun, M.; Yang, B.; Xiao, J.; Du, J. Ultrasound-Responsive Polymersomes Capable of Endosomal Escape for Efficient Cancer Therapy. J. Control. Release 2020, 322, 81–94. [Google Scholar] [CrossRef]
- Xu, J.; Yi, X.; Zhao, D.; Yuan, G.; Zhuo, R.; Li, F. A Three-Drug Co-Delivery System Based on Reduction-Sensitive Polymeric Prodrug to Effectively Reverse Multi-Drug Resistance. Chem. Res. Chin. Univ. 2017, 33, 484–491. [Google Scholar] [CrossRef]
- Yildirim, T.; Traeger, A.; Sungur, P.; Hoeppener, S.; Kellner, C.; Yildirim, I.; Pretzel, D.; Schubert, S.; Schubert, U.S. Polymersomes with Endosomal pH-Induced Vesicle-to-Micelle Morphology Transition and a Potential Application for Controlled Doxorubicin Delivery. Biomacromolecules 2017, 18, 3280–3290. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Dirisala, A.; Ge, Z.; Wang, Y.; Yin, W.; Ke, W.; Toh, K.; Xie, J.; Matsumoto, Y.; Anraku, Y.; et al. Therapeutic Vesicular Nanoreactors with Tumor-Specific Activation and Self-Destruction for Synergistic Tumor Ablation. Angew. Chem. Int. Ed. 2017, 56, 14025–14030. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Ge, Z.; Toh, K.; Liu, X.; Dirisala, A.; Ke, W.; Wen, P.; Zhou, H.; Wang, Z.; Xiao, S.; et al. Enzymatically Transformable Polymersome-Based Nanotherapeutics to Eliminate Minimal Relapsable Cancer. Adv. Mater. 2021, 33, 2105254. [Google Scholar] [CrossRef]
- Joglekar, M.; Trewyn, B.G. Polymer-Based Stimuli-Responsive Nanosystems for Biomedical Applications. Biotechnol. J. 2013, 8, 931–945. [Google Scholar] [CrossRef]
- Meng, F.; Zhong, Z.; Feijen, J. Stimuli-Responsive Polymersomes for Programmed Drug Delivery. Biomacromolecules 2009, 10, 197–209. [Google Scholar] [CrossRef]
- Anajafi, T.; Scott, M.D.; You, S.; Yang, X.; Choi, Y.; Qian, S.Y.; Mallik, S. Acridine Orange Conjugated Polymersomes for Simultaneous Nuclear Delivery of Gemcitabine and Doxorubicin to Pancreatic Cancer Cells. Bioconjug. Chem. 2016, 27, 762–771. [Google Scholar] [CrossRef]
- Karandish, F.; Mamnoon, B.; Feng, L.; Haldar, M.K.; Xia, L.; Gange, K.N.; You, S.; Choi, Y.; Sarkar, K.; Mallik, S. Nucleus-Targeted, Echogenic Polymersomes for Delivering a Cancer Stemness Inhibitor to Pancreatic Cancer Cells. Biomacromolecules 2018, 19, 4122–4132. [Google Scholar] [CrossRef]
- Nahire, R.; Haldar, M.K.; Paul, S.; Ambre, A.H.; Meghnani, V.; Layek, B.; Katti, K.S.; Gange, K.N.; Singh, J.; Sarkar, K.; et al. Multifunctional Polymersomes for Cytosolic Delivery of Gemcitabine and Doxorubicin to Cancer Cells. Biomaterials 2014, 35, 6482–6497. [Google Scholar] [CrossRef]
- Bej, R.; Ghosh, S. Glutathione Triggered Cascade Degradation of an Amphiphilic Poly(Disulfide)-Drug Conjugate and Targeted Release. Bioconjug. Chem. 2019, 30, 101–110. [Google Scholar] [CrossRef]
- Tsai, M.F.; Lo, Y.L.; Soorni, Y.; Su, C.H.; Sivasoorian, S.S.; Yang, J.Y.; Wang, L.F. Near-Infrared Light-Triggered Drug Release from Ultraviolet- And Redox-Responsive Polymersome Encapsulated with Core-Shell Upconversion Nanoparticles for Cancer Therapy. ACS Appl. Bio Mater. 2021, 4, 3264–3275. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.; Meng, F.; Deng, C.; Zhong, Z. Robust, Tumor-Homing and Redox-Sensitive Polymersomal Doxorubicin: A Superior Alternative to Doxil and Caelyx? J. Control. Release 2016, 239, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Benito, E.; Romero-Azogil, L.; Galbis, E.; De-Paz, M.V.; García-Martín, M.G. Structurally Simple Redox Polymersomes for Doxorubicin Delivery. Eur. Polym. J. 2020, 137, 109952. [Google Scholar] [CrossRef]
- Mohammadi, M.; Ramezani, M.; Abnous, K.; Alibolandi, M. Biocompatible Polymersomes-Based Cancer Theranostics: Towards Multifunctional Nanomedicine. Int. J. Pharm. 2017, 519, 287–303. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Chen, W.; Zheng, M.; Meng, F.; Zhong, Z. pH-Sensitive Degradable Chimaeric Polymersomes for the Intracellular Release of Doxorubicin Hydrochloride. Biomaterials 2012, 33, 7291–7299. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Hu, M.; Qiu, L. Drug Resistance Reversal by Combretastatin-A4 Phosphate Loaded with Doxorubicin in Polymersomes Independent of Angiogenesis Effect. J. Pharm. Pharmacol. 2017, 69, 844–855. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Meng, F.; Cheng, R.; Zhong, Z. pH-Sensitive Degradable Polymersomes for Triggered Release of Anticancer Drugs: A Comparative Study with Micelles. J. Control. Release 2010, 142, 40–46. [Google Scholar] [CrossRef]
- Guo, C.; Yuan, H.; Zhang, Y.; Yin, T.; He, H.; Gou, J.; Tang, X. Asymmetric Polymersomes, from the Formation of Asymmetric Membranes to the Application on Drug Delivery. J. Control. Release 2021, 338, 422–445. [Google Scholar] [CrossRef]
- Hu, M.; Shen, Y.; Zhang, L.; Qiu, L. Polymersomes via Self-Assembly of Amphiphilic β-Cyclodextrin-Centered Triarm Star Polymers for Enhanced Oral Bioavailability of Water-Soluble Chemotherapeutics. Biomacromolecules 2016, 17, 1026–1039. [Google Scholar] [CrossRef]
- Quadir, M.A.; Morton, S.W.; Mensah, L.B.; Shopsowitz, K.; Dobbelaar, J.; Effenberger, N.; Hammond, P.T. Ligand-Decorated Click Polypeptide Derived Nanoparticles for Targeted Drug Delivery Applications. Nanomed. Nanotechnol. Biol. Med. 2017, 13, 1797–1808. [Google Scholar] [CrossRef]
- Laskar, P.; Dey, J.; Ghosh, S.K. Spontaneously Formed Redox- and pH-Sensitive Polymersomes by MPEG Based Cytocompatible Random Copolymers. J. Colloid Interface Sci. 2017, 501, 22–33. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Liang, L.; Qiu, L. In Situ Generated Gold Nanoparticle Hybrid Polymersomes for Water-Soluble Chemotherapeutics: Inhibited Leakage and pH-Responsive Intracellular Release. Adv. Funct. Mater. 2017, 27, 1604981. [Google Scholar] [CrossRef]
- Liu, G.; Ma, S.; Li, S.; Cheng, R.; Meng, F.; Liu, H.; Zhong, Z. The Highly Efficient Delivery of Exogenous Proteins into Cells Mediated by Biodegradable Chimaeric Polymersomes. Biomaterials 2010, 31, 7575–7585. [Google Scholar] [CrossRef] [PubMed]
- Bartenstein, J.E.; Robertson, J.; Battaglia, G.; Briscoe, W.H. Stability of Polymersomes Prepared by Size Exclusion Chromatography and Extrusion. Colloids Surfaces A Physicochem. Eng. Asp. 2016, 506, 739–746. [Google Scholar] [CrossRef]
- Kumar, R.; Kulkarni, A.; Nagesha, D.K.; Sridhar, S. In Vitro Evaluation of Theranostic Polymeric Micelles for Imaging and Drug Delivery in Cancer. Theranostics 2012, 2, 714–722. [Google Scholar] [CrossRef] [PubMed]
- Mason, A.F.; Thordarson, P. Polymersomes with Asymmetric Membranes Based on Readily Accessible Di- and Triblock Copolymers Synthesized via SET-LRP. ACS Macro Lett. 2016, 5, 1172–1175. [Google Scholar] [CrossRef]
- Anajafi, T.; Yu, J.; Sedigh, A.; Haldar, M.K.; Muhonen, W.W.; Oberlander, S.; Wasness, H.; Froberg, J.; Molla, M.S.; Katti, K.S.; et al. Nuclear Localizing Peptide-Conjugated, Redox-Sensitive Polymersomes for Delivering Curcumin and Doxorubicin to Pancreatic Cancer Microtumors. Mol. Pharm. 2017, 14, 1916–1928. [Google Scholar] [CrossRef]
- Luo, Z.; Li, Y.; Wang, B.; Jiang, J. pH-Sensitive Vesicles Formed by Amphiphilic Grafted Copolymers with Tunable Membrane Permeability for Drug Loading/Release: A Multiscale Simulation Study. Macromolecules 2016, 49, 6084–6094. [Google Scholar] [CrossRef]
- Liu, H.; Gong, L.; Lu, S.; Wang, H.; Fan, W.; Yang, C. Three Core-Shell Polymersomes for Targeted Doxorubicin Delivery: Sustained and Acidic Release. J. Drug Deliv. Sci. Technol. 2021, 61, 102293. [Google Scholar] [CrossRef]
- Shen, J.; Burgess, D.J. In Vitro Dissolution Testing Strategies for Nanoparticulate Drug Delivery Systems: Recent Developments and Challenges. Drug Deliv. Transl. Res. 2013, 3, 409–415. [Google Scholar] [CrossRef]
- Wallace, S.J.; Li, J.; Nation, R.L.; Boyd, B.J. Drug Release from Nanomedicines: Selection of Appropriate Encapsulation and Release Methodology. Drug Deliv. Transl. Res. 2012, 2, 284–292. [Google Scholar] [CrossRef] [PubMed]
- De Paz, M.V.; Zamora, F.; Begines, B.; Ferris, C.; Galbis, J.A. Glutathione-Mediated Biodegradable Polyurethanes Derived from L-Arabinitol. Biomacromolecules 2010, 11, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Ferris, C.; De Paz, M.V.; Aguilar-De-Leyva, Á.; Caraballo, I.; Galbis, J.A. Reduction-Sensitive Functionalized Copolyurethanes for Biomedical Applications. Polym. Chem. 2014, 5, 2370–2381. [Google Scholar] [CrossRef]
- Bej, R.; Sarkar, J.; Ray, D.; Aswal, V.K.; Ghosh, S. Morphology Regulation in Redox Destructible Amphiphilic Block Copolymers and Impact on Intracellular Drug Delivery. Macromol. Biosci. 2018, 18, 1800057. [Google Scholar] [CrossRef]
- Costa, P.; Sousa Lobo, J.M. Modeling and Comparison of Dissolution Profiles. Eur. J. Pharm. Sci. 2001, 13, 123–133. [Google Scholar] [CrossRef]
- Peppas, N.A.; Sahlin, J.J. A Simple Equation for the Description of Solute Release. III. Coupling of Diffusion and Relaxation. Int. J. Pharm. 1989, 57, 169–172. [Google Scholar] [CrossRef]










| Dox·HCl (Theoretical Load) | Average Diameter (nm) | PDI | Zeta Potential (mV) | DEE (wt.%) | DLC (wt.%) |
|---|---|---|---|---|---|
| 5 wt.% | 139.1 ± 7.5 | 0.151 ± 0.016 | −7.0 ± 1.5 | 93.83 ± 0.01 | 4.69 ± 0.00 |
| 10 wt.% | 124.2 ± 12.0 | 0.164 ± 0.071 | −23.8 ± 0.9 | 98.26 ± 0.10 | 9.83 ± 0.01 |
| Kinetic Model | Parameters | pH 7.4 Release Medium | pH 7.4/50 mM GSH Release Medium |
|---|---|---|---|
| Zero-order (3) | k0 (h−1) r2adj | 0.0049 0.8617 (F = 70) | 0.0146 0.8074 (F = 26) |
| First-order (4) | k1 (h−1) r2adj | 0.0065 0.8977 (F = 98) | 0.0297 0.9348 (F = 87) |
| Higuchi (5) | kH (h−0.5) r2adj | 0.0392 0.9572 (F = 247) | 0.1230 0.9154 (F = 66) |
| Korsmeyer–Peppas (6) | n kK (h−n) r2adj | 0.31 1.1077 0.9762 (F = 452) | 0.76 1.0549 0.8466 (F = 34) |
| Peppas–Sahlin (7) | kd (h−0.43) kr (h−0.86) r2adj | 0.0827 −0.0048 0.9755 (F = 220) | 0.2489 −0.0125 0.9174 (F = 34) |
| Hixson–Crowell (8) | kHC (h−1) r2adj | 0.0020 0.8862 (F = 87) | 0.0077 0.8993 (F = 55) |
| Baker–Lonsdale (9) | kBL (h−1) r2adj | 0.0005 0.9700 (F = 356) | 0.0036 0.9829 (F = 347) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferrero, C.; Casas, M.; Caraballo, I. Redox-Responsive Polymersomes as Smart Doxorubicin Delivery Systems. Pharmaceutics 2022, 14, 1724. https://doi.org/10.3390/pharmaceutics14081724
Ferrero C, Casas M, Caraballo I. Redox-Responsive Polymersomes as Smart Doxorubicin Delivery Systems. Pharmaceutics. 2022; 14(8):1724. https://doi.org/10.3390/pharmaceutics14081724
Chicago/Turabian StyleFerrero, Carmen, Marta Casas, and Isidoro Caraballo. 2022. "Redox-Responsive Polymersomes as Smart Doxorubicin Delivery Systems" Pharmaceutics 14, no. 8: 1724. https://doi.org/10.3390/pharmaceutics14081724
APA StyleFerrero, C., Casas, M., & Caraballo, I. (2022). Redox-Responsive Polymersomes as Smart Doxorubicin Delivery Systems. Pharmaceutics, 14(8), 1724. https://doi.org/10.3390/pharmaceutics14081724