Uptake Transporters at the Blood–Brain Barrier and Their Role in Brain Drug Disposition
Abstract
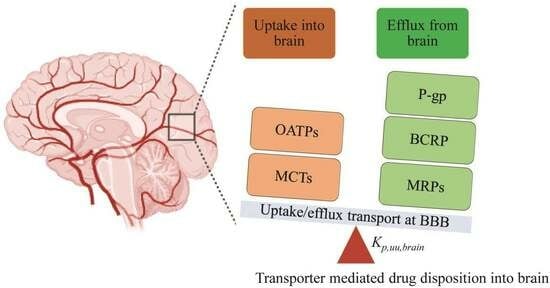
:1. Introduction
2. Study Highlights
3. Localization, Functions, and Expression of the Drug Uptake Transporters in the Brain
3.1. Models Used to Study Uptake Transporters-Mediated Brain Drug Disposition
3.2. Species Differences in Uptake Transporter Activity and Drug–Drug Interactions in Brain
3.3. Significance of Uptake Transporters in Brain Drug Disposition
4. Conclusions and Future Directions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Giacomini, K.M.; Huang, S.-M.; Tweedie, D.J.; Benet, L.Z.; Brouwer, K.L.R.; Chu, X.; Dahlin, A.; Evers, R.; Fischer, V.; Hillgren, K.M.; et al. Membrane Transporters in Drug Development. Nat. Rev. Drug Discov. 2010, 9, 215–236. [Google Scholar] [CrossRef]
- Alavijeh, M.S.; Chishty, M.; Qaiser, M.Z.; Palmer, A.M. Drug Metabolism and Pharmacokinetics, the Blood-Brain Barrier, and Central Nervous System Drug Discovery. NeuroRx 2005, 2, 554–571. [Google Scholar] [CrossRef] [PubMed]
- Osborne, O.; Peyravian, N.; Nair, M.; Daunert, S.; Toborek, M. The Paradox of HIV Blood-Brain Barrier Penetrance and Antiretroviral Drug Delivery Deficiencies. Trends Neurosci. 2020, 43, 695–708. [Google Scholar] [CrossRef] [PubMed]
- Loryan, I.; Reichel, A.; Feng, B.; Bundgaard, C.; Shaffer, C.; Kalvass, C.; Bednarczyk, D.; Morrison, D.; Lesuisse, D.; Hoppe, E.; et al. Unbound Brain-to-Plasma Partition Coefficient, Kp,Uu,Brain—A Game Changing Parameter for CNS Drug Discovery and Development. Pharm. Res. 2022, 39, 1321–1341. [Google Scholar] [CrossRef]
- Hoque, M.T.; Kis, O.; Rosa, M.F.D.; Bendayan, R. Raltegravir Permeability across Blood-Tissue Barriers and the Potential Role of Drug Efflux Transporters. Antimicrob. Agents Chemother. 2015, 59, 2572–2582. [Google Scholar] [CrossRef]
- Al-Majdoub, Z.M.; Feteisi, H.A.; Achour, B.; Warwood, S.; Neuhoff, S.; Rostami-Hodjegan, A.; Barber, J. Proteomic Quantification of Human Blood-Brain Barrier SLC and ABC Transporters in Healthy Individuals and Dementia Patients. Mol. Pharm. 2019, 16, 1220–1233. [Google Scholar] [CrossRef] [PubMed]
- Ohtsuki, S.; Ikeda, C.; Uchida, Y.; Sakamoto, Y.; Miller, F.; Glacial, F.; Decleves, X.; Scherrmann, J.-M.; Couraud, P.-O.; Kubo, Y.; et al. Quantitative Targeted Absolute Proteomic Analysis of Transporters, Receptors and Junction Proteins for Validation of Human Cerebral Microvascular Endothelial Cell Line HCMEC/D3 as a Human Blood-Brain Barrier Model. Mol. Pharm. 2013, 10, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Uchida, Y.; Ohtsuki, S.; Katsukura, Y.; Ikeda, C.; Suzuki, T.; Kamiie, J.; Terasaki, T. Quantitative Targeted Absolute Proteomics of Human Blood-Brain Barrier Transporters and Receptors. J. Neurochem. 2011, 117, 333–345. [Google Scholar] [CrossRef]
- Uchida, Y.; Zhang, Z.; Tachikawa, M.; Terasaki, T. Quantitative Targeted Absolute Proteomics of Rat Blood-Cerebrospinal Fluid Barrier Transporters: Comparison with a Human Specimen. J. Neurochem. 2015, 134, 1104–1115. [Google Scholar] [CrossRef] [PubMed]
- Bao, X.; Wu, J.; Xie, Y.; Kim, S.; Michelhaugh, S.; Jiang, J.; Mittal, S.; Sanai, N.; Li, J. Protein Expression and Functional Relevance of Efflux and Uptake Drug Transporters at the Blood-Brain Barrier of Human Brain and Glioblastoma. Clin. Pharmacol. Ther. 2020, 107, 1116–1127. [Google Scholar] [CrossRef]
- Su, Y.; Zhang, X.; Sinko, P.J. Human Organic Anion-Transporting Polypeptide OATP-A (SLC21A3) Acts in Concert with P-Glycoprotein and Multidrug Resistance Protein 2 in the Vectorial Transport of Saquinavir in Hep G2 Cells. Mol. Pharm. 2004, 1, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Maeda, T.; Takahashi, K.; Ohtsu, N.; Oguma, T.; Ohnishi, T.; Atsumi, R.; Tamai, I. Identification of Influx Transporter for the Quinolone Antibacterial Agent Levofloxacin. Mol. Pharm. 2007, 4, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Franke, R.M.; Baker, S.D.; Mathijssen, R.H.; Schuetz, E.G.; Sparreboom, A. Influence of Solute Carriers on the Pharmacokinetics of CYP3A4 Probes. Clin. Pharmacol. Ther. 2008, 84, 704–709. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Franke, R.M.; Filipski, K.K.; Hu, C.; Orwick, S.J.; de Bruijn, E.A.; Burger, H.; Baker, S.D.; Sparreboom, A. Interaction of Imatinib with Human Organic Ion Carriers. Clin. Cancer Res. 2008, 14, 3141–3148. [Google Scholar] [CrossRef]
- Cvetkovic, M.; Leake, B.; Fromm, M.F.; Wilkinson, G.R.; Kim, R.B. OATP and P-Glycoprotein Transporters Mediate the Cellular Uptake and Excretion of Fexofenadine. Drug Metab. Dispos. 1999, 27, 866–871. [Google Scholar]
- Fujino, H.; Saito, T.; Ogawa, S.-I.; Kojima, J. Transporter-Mediated Influx and Efflux Mechanisms of Pitavastatin, a New Inhibitor of HMG-CoA Reductase. J. Pharm. Pharmacol. 2005, 57, 1305–1311. [Google Scholar] [CrossRef]
- Treiber, A.; Schneiter, R.; Häusler, S.; Stieger, B. Bosentan Is a Substrate of Human OATP1B1 and OATP1B3: Inhibition of Hepatic Uptake as the Common Mechanism of Its Interactions with Cyclosporin A, Rifampicin, and Sildenafil. Drug Metab. Dispos. 2007, 35, 1400–1407. [Google Scholar] [CrossRef]
- Satoh, H.; Yamashita, F.; Tsujimoto, M.; Murakami, H.; Koyabu, N.; Ohtani, H.; Sawada, Y. Citrus Juices Inhibit the Function of Human Organic Anion-Transporting Polypeptide OATP-B. Drug Metab. Dispos. 2005, 33, 518–523. [Google Scholar] [CrossRef]
- Ho, R.H.; Tirona, R.G.; Leake, B.F.; Glaeser, H.; Lee, W.; Lemke, C.J.; Wang, Y.; Kim, R.B. Drug and Bile Acid Transporters in Rosuvastatin Hepatic Uptake: Function, Expression, and Pharmacogenetics. Gastroenterology 2006, 130, 1793–1806. [Google Scholar] [CrossRef]
- Nozawa, T.; Imai, K.; Nezu, J.-I.; Tsuji, A.; Tamai, I. Functional Characterization of PH-Sensitive Organic Anion Transporting Polypeptide OATP-B in Human. J. Pharmacol. Exp. Ther. 2004, 308, 438–445. [Google Scholar] [CrossRef]
- Tamai, I.; Nezu, J.; Uchino, H.; Sai, Y.; Oku, A.; Shimane, M.; Tsuji, A. Molecular Identification and Characterization of Novel Members of the Human Organic Anion Transporter (OATP) Family. Biochem. Biophys. Res. Commun. 2000, 273, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Grube, M.; Köck, K.; Oswald, S.; Draber, K.; Meissner, K.; Eckel, L.; Böhm, M.; Felix, S.B.; Vogelgesang, S.; Jedlitschky, G.; et al. Organic Anion Transporting Polypeptide 2B1 Is a High-Affinity Transporter for Atorvastatin and Is Expressed in the Human Heart. Clin. Pharmacol. Ther. 2006, 80, 607–620. [Google Scholar] [CrossRef]
- Kullak-Ublick, G.A.; Fisch, T.; Oswald, M.; Hagenbuch, B.; Meier, P.J.; Beuers, U.; Paumgartner, G. Dehydroepiandrosterone Sulfate (DHEAS): Identification of a Carrier Protein in Human Liver and Brain. FEBS Lett. 1998, 424, 173–176. [Google Scholar] [CrossRef]
- Dresser, G.K.; Bailey, D.G.; Leake, B.F.; Schwarz, U.I.; Dawson, P.A.; Freeman, D.J.; Kim, R.B. Fruit Juices Inhibit Organic Anion Transporting Polypeptide-Mediated Drug Uptake to Decrease the Oral Availability of Fexofenadine. Clin. Pharmacol. Ther. 2002, 71, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Bailey, D.G.; Dresser, G.K.; Leake, B.F.; Kim, R.B. Naringin Is a Major and Selective Clinical Inhibitor of Organic Anion-Transporting Polypeptide 1A2 (OATP1A2) in Grapefruit Juice. Clin. Pharmacol. Ther. 2007, 81, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Zhao, R.; Kalvass, J.C.; Yanni, S.B.; Bridges, A.S.; Pollack, G.M. Fexofenadine Brain Exposure and the Influence of Blood-Brain Barrier P-Glycoprotein after Fexofenadine and Terfenadine Administration. Drug Metab. Dispos. 2009, 37, 529–535. [Google Scholar] [CrossRef] [PubMed]
- Neuhaus, W.; Mandikova, J.; Pawlowitsch, R.; Linz, B.; Bennani-Baiti, B.; Lauer, R.; Lachmann, B.; Noe, C.R. Blood-Brain Barrier In Vitro Models as Tools in Drug Discovery: Assessment of the Transport Ranking of Antihistaminic Drugs. Pharmazie 2012, 67, 432–439. [Google Scholar] [PubMed]
- Kalliokoski, A.; Niemi, M. Impact of OATP Transporters on Pharmacokinetics. Br. J. Pharmacol. 2009, 158, 693–705. [Google Scholar] [CrossRef]
- Tahara, H.; Kusuhara, H.; Fuse, E.; Sugiyama, Y. P-Glycoprotein Plays a Major Role in the Efflux of Fexofenadine in the Small Intestine and Blood-Brain Barrier, but Only a Limited Role in Its Biliary Excretion. Drug Metab. Dispos. 2005, 33, 963–968. [Google Scholar] [CrossRef]
- Chan, G.N.Y.; Hoque, M.T.; Bendayan, R. Role of Nuclear Receptors in the Regulation of Drug Transporters in the Brain. Trends Pharmacol. Sci. 2013, 34, 361–372. [Google Scholar] [CrossRef]
- Lin, C.-J.; Tai, Y.; Huang, M.-T.; Tsai, Y.-F.; Hsu, H.-J.; Tzen, K.-Y.; Liou, H.-H. Cellular Localization of the Organic Cation Transporters, OCT1 and OCT2, in Brain Microvessel Endothelial Cells and Its Implication for MPTP Transport across the Blood-Brain Barrier and MPTP-Induced Dopaminergic Toxicity in Rodents. J. Neurochem. 2010, 114, 717–727. [Google Scholar] [CrossRef]
- Geier, E.G.; Chen, E.C.; Webb, A.; Papp, A.C.; Yee, S.W.; Sadee, W.; Giacomini, K.M. Profiling Solute Carrier Transporters in the Human Blood-Brain Barrier. Clin. Pharmacol Ther. 2013, 94, 636–639. [Google Scholar] [CrossRef]
- Friedrich, A.; Prasad, P.D.; Freyer, D.; Ganapathy, V.; Brust, P. Molecular Cloning and Functional Characterization of the OCTN2 Transporter at the RBE4 Cells, an In Vitro Model of the Blood-Brain Barrier. Brain Res. 2003, 968, 69–79. [Google Scholar] [CrossRef]
- Morris, M.E.; Rodriguez-Cruz, V.; Felmlee, M.A. SLC and ABC Transporters: Expression, Localization, and Species Differences at the Blood-Brain and the Blood-Cerebrospinal Fluid Barriers. AAPS J. 2017, 19, 1317–1331. [Google Scholar] [CrossRef] [PubMed]
- Halestrap, A.P.; Meredith, D. The SLC16 Gene Family-from Monocarboxylate Transporters (MCTs) to Aromatic Amino Acid Transporters and Beyond. Pflugers Arch. 2004, 447, 619–628. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, Y.; Bai, Y.; Dai, L.; Guo, H. Monocarboxylate Transporter 1 May Benefit Cerebral Ischemia via Facilitating Lactate Transport From Glial Cells to Neurons. Front. Neurol. 2022, 13, 781063. [Google Scholar] [CrossRef] [PubMed]
- Vijay, N.; Morris, M.E. Role of Monocarboxylate Transporters in Drug Delivery to the Brain. Curr. Pharm. Des. 2014, 20, 1487–1498. [Google Scholar] [CrossRef] [PubMed]
- Scalise, M.; Galluccio, M.; Console, L.; Pochini, L.; Indiveri, C. The Human SLC7A5 (LAT1): The Intriguing Histidine/Large Neutral Amino Acid Transporter and Its Relevance to Human Health. Front. Chem. 2018, 6, 243. [Google Scholar] [CrossRef]
- Friesema, E.C.; Docter, R.; Moerings, E.P.; Verrey, F.; Krenning, E.P.; Hennemann, G.; Visser, T.J. Thyroid Hormone Transport by the Heterodimeric Human System L Amino Acid Transporter. Endocrinology 2001, 142, 4339–4348. [Google Scholar] [CrossRef]
- del Amo, E.M.; Urtti, A.; Yliperttula, M. Pharmacokinetic Role of L-Type Amino Acid Transporters LAT1 and LAT2. Eur. J. Pharm. Sci. 2008, 35, 161–174. [Google Scholar] [CrossRef]
- Murata, Y.; Neuhoff, S.; Rostami-Hodjegan, A.; Takita, H.; Al-Majdoub, Z.M.; Ogungbenro, K. In Vitro to In Vivo Extrapolation Linked to Physiologically Based Pharmacokinetic Models for Assessing the Brain Drug Disposition. AAPS J. 2022, 24, 28. [Google Scholar] [CrossRef]
- Puris, E.; Fricker, G.; Gynther, M. Targeting Transporters for Drug Delivery to the Brain: Can We Do Better? Pharm. Res. 2022, 39, 1415–1455. [Google Scholar] [CrossRef]
- Rihani, S.B.A.; Darakjian, L.I.; Deodhar, M.; Dow, P.; Turgeon, J.; Michaud, V. Disease-Induced Modulation of Drug Transporters at the Blood–Brain Barrier Level. Int. J. Mol. Sci. 2021, 22, 3742. [Google Scholar] [CrossRef]
- Sanchez-Covarrubias, L.; Slosky, L.M.; Thompson, B.J.; Davis, T.P.; Ronaldson, P.T. Transporters at CNS Barrier Sites: Obstacles or Opportunities for Drug Delivery? Curr. Pharm. Des. 2014, 20, 1422–1449. [Google Scholar] [CrossRef]
- Lee, W.; Glaeser, H.; Smith, L.H.; Roberts, R.L.; Moeckel, G.W.; Gervasini, G.; Leake, B.F.; Kim, R.B. Polymorphisms in Human Organic Anion-Transporting Polypeptide 1A2 (OATP1A2): Implications for Altered Drug Disposition and Central Nervous System Drug Entry. J. Biol. Chem. 2005, 280, 9610–9617. [Google Scholar] [CrossRef]
- Gao, B.; Hagenbuch, B.; Kullak-Ublick, G.A.; Benke, D.; Aguzzi, A.; Meier, P.J. Organic Anion-Transporting Polypeptides Mediate Transport of Opioid Peptides across Blood-Brain Barrier. J. Pharmacol. Exp. Ther. 2000, 294, 73–79. [Google Scholar] [PubMed]
- Bronger, H.; König, J.; Kopplow, K.; Steiner, H.-H.; Ahmadi, R.; Herold-Mende, C.; Keppler, D.; Nies, A.T. ABCC Drug Efflux Pumps and Organic Anion Uptake Transporters in Human Gliomas and the Blood-Tumor Barrier. Cancer Res. 2005, 65, 11419–11428. [Google Scholar] [CrossRef] [PubMed]
- Betterton, R.D.; Davis, T.P.; Ronaldson, P.T. Organic Cation Transporters in the Central Nervous System. Handb. Exp. Pharmacol. 2021, 266, 301–328. [Google Scholar] [CrossRef]
- Billington, S.; Salphati, L.; Hop, C.E.C.A.; Chu, X.; Evers, R.; Burdette, D.; Rowbottom, C.; Lai, Y.; Xiao, G.; Humphreys, W.G.; et al. Interindividual and Regional Variability in Drug Transporter Abundance at the Human Blood-Brain Barrier Measured by Quantitative Targeted Proteomics. Clin. Pharmacol. Ther. 2019, 106, 228–237. [Google Scholar] [CrossRef]
- Bay, C.; Bajraktari-Sylejmani, G.; Haefeli, W.E.; Burhenne, J.; Weiss, J.; Sauter, M. Functional Characterization of the Solute Carrier LAT-1 (SLC7A5/SLC2A3) in Human Brain Capillary Endothelial Cells with Rapid UPLC-MS/MS Quantification of Intracellular Isotopically Labelled L-Leucine. Int. J. Mol. Sci. 2022, 23, 3637. [Google Scholar] [CrossRef] [PubMed]
- Yan, R.; Zhao, X.; Lei, J.; Zhou, Q. Structure of the Human LAT1-4F2hc Heteromeric Amino Acid Transporter Complex. Nature 2019, 568, 127–130. [Google Scholar] [CrossRef]
- Haining, Z.; Kawai, N.; Miyake, K.; Okada, M.; Okubo, S.; Zhang, X.; Fei, Z.; Tamiya, T. Relation of LAT1/4F2hc Expression with Pathological Grade, Proliferation and Angiogenesis in Human Gliomas. BMC Clin. Pathol. 2012, 12, 4. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, T.; Sanada, Y.; Hattori, Y.; Suzuki, M. Correlation between the Expression of LAT1 in Cancer Cells and the Potential Efficacy of Boron Neutron Capture Therapy. J. Radiat. Res. 2023, 64, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Bauer, M.; Matsuda, A.; Wulkersdorfer, B.; Philippe, C.; Traxl, A.; Özvegy-Laczka, C.; Stanek, J.; Nics, L.; Klebermass, E.-M.; Poschner, S.; et al. Influence of OATPs on Hepatic Disposition of Erlotinib Measured With Positron Emission Tomography. Clin. Pharmacol. Ther. 2018, 104, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Wolburg, H.; Lippoldt, A. Tight Junctions of the Blood-Brain Barrier: Development, Composition and Regulation. Vascul. Pharmacol. 2002, 38, 323–337. [Google Scholar] [CrossRef]
- Helms, H.C.; Abbott, N.J.; Burek, M.; Cecchelli, R.; Couraud, P.-O.; Deli, M.A.; Förster, C.; Galla, H.J.; Romero, I.A.; Shusta, E.V.; et al. In Vitro Models of the Blood-Brain Barrier: An Overview of Commonly Used Brain Endothelial Cell Culture Models and Guidelines for Their Use. J. Cereb. Blood Flow Metab. 2016, 36, 862–890. [Google Scholar] [CrossRef]
- Vallianatou, T.; Tsopelas, F.; Tsantili-Kakoulidou, A. Prediction Models for Brain Distribution of Drugs Based on Biomimetic Chromatographic Data. Molecules 2022, 27, 3668. [Google Scholar] [CrossRef]
- Joó, F.; Karnushina, I. A Procedure for the Isolation of Capillaries from Rat Brain. Cytobios 1973, 8, 41–48. [Google Scholar] [PubMed]
- Erdlenbruch, B.; Alipour, M.; Fricker, G.; Miller, D.S.; Kugler, W.; Eibl, H.; Lakomek, M. Alkylglycerol Opening of the Blood-Brain Barrier to Small and Large Fluorescence Markers in Normal and C6 Glioma-Bearing Rats and Isolated Rat Brain Capillaries. Br. J. Pharmacol. 2003, 140, 1201–1210. [Google Scholar] [CrossRef]
- DeBault, L.E.; Kahn, L.E.; Frommes, S.P.; Cancilla, P.A. Cerebral Microvessels and Derived Cells in Tissue Culture: Isolation and Preliminary Characterization. In Vitro 1979, 15, 473–487. [Google Scholar] [CrossRef]
- Cecchelli, R.; Dehouck, B.; Descamps, L.; Fenart, L.; Buée-Scherrer, V.V.; Duhem, C.; Lundquist, S.; Rentfel, M.; Torpier, G.; Dehouck, M. In Vitro Model for Evaluating Drug Transport across the Blood-Brain Barrier. Adv. Drug Deliv. Rev. 1999, 36, 165–178. [Google Scholar] [CrossRef]
- Dehouck, M.P.; Méresse, S.; Delorme, P.; Fruchart, J.C.; Cecchelli, R. An Easier, Reproducible, and Mass-Production Method to Study the Blood-Brain Barrier In Vitro. J. Neurochem. 1990, 54, 1798–1801. [Google Scholar] [CrossRef] [PubMed]
- Gaillard, P.J.; Voorwinden, L.H.; Nielsen, J.L.; Ivanov, A.; Atsumi, R.; Engman, H.; Ringbom, C.; de Boer, A.G.; Breimer, D.D. Establishment and Functional Characterization of an In Vitro Model of the Blood-Brain Barrier, Comprising a Co-Culture of Brain Capillary Endothelial Cells and Astrocytes. Eur. J. Pharm. Sci. 2001, 12, 215–222. [Google Scholar] [CrossRef]
- DeBault, L.E.; Cancilla, P.A. Gamma-Glutamyl Transpeptidase in Isolated Brain Endothelial Cells: Induction by Glial Cells In Vitro. Science 1980, 207, 653–655. [Google Scholar] [CrossRef] [PubMed]
- Tao-Cheng, J.H.; Nagy, Z.; Brightman, M.W. Tight Junctions of Brain Endothelium In Vitro Are Enhanced by Astroglia. J. Neurosci. 1987, 7, 3293–3299. [Google Scholar] [CrossRef] [PubMed]
- Thomsen, L.B.; Burkhart, A.; Moos, T. A Triple Culture Model of the Blood-Brain Barrier Using Porcine Brain Endothelial Cells, Astrocytes and Pericytes. PLoS ONE 2015, 10, e0134765. [Google Scholar] [CrossRef]
- Nakagawa, S.; Deli, M.A.; Kawaguchi, H.; Shimizudani, T.; Shimono, T.; Kittel, A.; Tanaka, K.; Niwa, M. A New Blood-Brain Barrier Model Using Primary Rat Brain Endothelial Cells, Pericytes and Astrocytes. Neurochem. Int. 2009, 54, 253–263. [Google Scholar] [CrossRef]
- Burek, M.; Förster, C.Y. Cloning and Characterization of the Murine Claudin-5 Promoter. Mol. Cell. Endocrinol. 2009, 298, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Blecharz, K.G.; Drenckhahn, D.; Förster, C.Y. Glucocorticoids Increase VE-Cadherin Expression and Cause Cytoskeletal Rearrangements in Murine Brain Endothelial CEND Cells. J. Cereb. Blood Flow Metab. 2008, 28, 1139–1149. [Google Scholar] [CrossRef] [PubMed]
- Harke, N.; Leers, J.; Kietz, S.; Drenckhahn, D.; Förster, C. Glucocorticoids Regulate the Human Occludin Gene through a Single Imperfect Palindromic Glucocorticoid Response Element. Mol. Cell. Endocrinol. 2008, 295, 39–47. [Google Scholar] [CrossRef]
- Förster, C.; Silwedel, C.; Golenhofen, N.; Burek, M.; Kietz, S.; Mankertz, J.; Drenckhahn, D. Occludin as Direct Target for Glucocorticoid-Induced Improvement of Blood-Brain Barrier Properties in a Murine In Vitro System. J. Physiol. 2005, 565, 475–486. [Google Scholar] [CrossRef]
- Hughes, C.C.; Lantos, P.L. Uptake of Leucine and Alanine by Cultured Cerebral Capillary Endothelial Cells. Brain Res. 1989, 480, 126–132. [Google Scholar] [CrossRef]
- Pifferi, F.; Jouin, M.; Alessandri, J.M.; Haedke, U.; Roux, F.; Perrière, N.; Denis, I.; Lavialle, M.; Guesnet, P. n-3 Fatty Acids Modulate Brain Glucose Transport in Endothelial Cells of the Blood-Brain Barrier. Prostaglandins Leukot. Essent. Fat. Acids 2007, 77, 279–286. [Google Scholar] [CrossRef] [PubMed]
- Pifferi, F.; Jouin, M.; Alessandri, J.-M.; Roux, F.; Perrière, N.; Langelier, B.; Lavialle, M.; Cunnane, S.; Guesnet, P. n-3 Long-Chain Fatty Acids and Regulation of Glucose Transport in Two Models of Rat Brain Endothelial Cells. Neurochem. Int. 2010, 56, 703–710. [Google Scholar] [CrossRef]
- Calabria, A.R.; Shusta, E.V. A Genomic Comparison of In Vivo and In Vitro Brain Microvascular Endothelial Cells. J. Cereb. Blood Flow Metab. 2008, 28, 135–148. [Google Scholar] [CrossRef] [PubMed]
- Garberg, P.; Ball, M.; Borg, N.; Cecchelli, R.; Fenart, L.; Hurst, R.D.; Lindmark, T.; Mabondzo, A.; Nilsson, J.E.; Raub, T.J.; et al. In Vitro Models for the Blood-Brain Barrier. Toxicol Vitr. 2005, 19, 299–334. [Google Scholar] [CrossRef]
- Weksler, B.; Romero, I.A.; Couraud, P.-O. The HCMEC/D3 Cell Line as a Model of the Human Blood Brain Barrier. Fluids Barriers CNS 2013, 10, 16. [Google Scholar] [CrossRef]
- Gromnicova, R.; Davies, H.A.; Sreekanthreddy, P.; Romero, I.A.; Lund, T.; Roitt, I.M.; Phillips, J.B.; Male, D.K. Glucose-Coated Gold Nanoparticles Transfer across Human Brain Endothelium and Enter Astrocytes In Vitro. PLoS ONE 2013, 8, e81043. [Google Scholar] [CrossRef] [PubMed]
- Steiner, O.; Coisne, C.; Engelhardt, B.; Lyck, R. Comparison of Immortalized BEnd5 and Primary Mouse Brain Microvascular Endothelial Cells as In Vitro Blood-Brain Barrier Models for the Study of T Cell Extravasation. J. Cereb. Blood Flow Metab. 2011, 31, 315–327. [Google Scholar] [CrossRef]
- Carl, S.M.; Lindley, D.J.; Das, D.; Couraud, P.O.; Weksler, B.B.; Romero, I.; Mowery, S.A.; Knipp, G.T. ABC and SLC Transporter Expression and Proton Oligopeptide Transporter (POT) Mediated Permeation across the Human Blood--Brain Barrier Cell Line, HCMEC/D3 [Corrected]. Mol. Pharm. 2010, 7, 1057–1068. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Ramirez, M.A.; Male, D.K.; Wang, C.; Sharrack, B.; Wu, D.; Romero, I.A. Cytokine-Induced Changes in the Gene Expression Profile of a Human Cerebral Microvascular Endothelial Cell-Line, HCMEC/D3. Fluids Barriers CNS 2013, 10, 27. [Google Scholar] [CrossRef]
- Stanton, J.A.; Williams, E.I.; Betterton, R.D.; Davis, T.P.; Ronaldson, P.T. Targeting Organic Cation Transporters at the Blood-Brain Barrier to Treat Ischemic Stroke in Rats. Exp. Neurol. 2022, 357, 114181. [Google Scholar] [CrossRef]
- Betterton, R.D.; Abdullahi, W.; Williams, E.I.; Lochhead, J.J.; Brzica, H.; Stanton, J.; Reddell, E.; Ogbonnaya, C.; Davis, T.P.; Ronaldson, P.T. Regulation of Blood-Brain Barrier Transporters by Transforming Growth Factor-β/Activin Receptor-Like Kinase 1 Signaling: Relevance to the Brain Disposition of 3-Hydroxy-3-Methylglutaryl Coenzyme A Reductase Inhibitors (i.e., Statins). Drug Metab. Dispos. 2022, 50, 942–956. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Chen, S.; Jia, J.; Liu, C.; Wang, W.; Zhang, H.; Zhen, X. Development and Evaluation of Novel Metformin Derivative Metformin Threonate for Brain Ischemia Treatment. Front. Pharmacol. 2022, 13, 879690. [Google Scholar] [CrossRef]
- Taskar, K.S.; Mariappan, T.T.; Kurawattimath, V.; Gautam, S.S.; Mullapudi, T.V.R.; Sridhar, S.K.; Kallem, R.R.; Marathe, P.; Mandlekar, S. Unmasking the Role of Uptake Transporters for Digoxin Uptake Across the Barriers of the Central Nervous System in Rat. J. Cent. Nerv. Syst. Dis. 2017, 9, 1179573517693596. [Google Scholar] [CrossRef]
- Marie, S.; Breuil, L.; Chalampalakis, Z.; Becquemont, L.; Verstuyft, C.; Lecoq, A.-L.; Caillé, F.; Gervais, P.; Lebon, V.; Comtat, C.; et al. [(11)C]Glyburide PET Imaging for Quantitative Determination of the Importance of Organic Anion-Transporting Polypeptide Transporter Function in the Human Liver and Whole-Body. Biomed. Pharmacother. 2022, 156, 113994. [Google Scholar] [CrossRef] [PubMed]
- Tournier, N.; Saba, W.; Cisternino, S.; Peyronneau, M.-A.; Damont, A.; Goutal, S.; Dubois, A.; Dollé, F.; Scherrmann, J.-M.; Valette, H.; et al. Effects of Selected OATP and/or ABC Transporter Inhibitors on the Brain and Whole-Body Distribution of Glyburide. AAPS J. 2013, 15, 1082–1090. [Google Scholar] [CrossRef]
- Ose, A.; Kusuhara, H.; Endo, C.; Tohyama, K.; Miyajima, M.; Kitamura, S.; Sugiyama, Y. Functional Characterization of Mouse Organic Anion Transporting Peptide 1a4 in the Uptake and Efflux of Drugs across the Blood-Brain Barrier. Drug Metab. Dispos. 2010, 38, 168–176. [Google Scholar] [CrossRef]
- Abdullahi, W.; Brzica, H.; Hirsch, N.A.; Reilly, B.G.; Ronaldson, P.T. Functional Expression of Organic Anion Transporting Polypeptide 1a4 Is Regulated by Transforming Growth Factor-β/Activin Receptor-like Kinase 1 Signaling at the Blood-Brain Barrier. Mol. Pharmacol. 2018, 94, 1321–1333. [Google Scholar] [CrossRef]
- Braun, C.; Sakamoto, A.; Fuchs, H.; Ishiguro, N.; Suzuki, S.; Cui, Y.; Klinder, K.; Watanabe, M.; Terasaki, T.; Sauer, A. Quantification of Transporter and Receptor Proteins in Dog Brain Capillaries and Choroid Plexus: Relevance for the Distribution in Brain and CSF of Selected BCRP and P-Gp Substrates. Mol. Pharm. 2017, 14, 3436–3447. [Google Scholar] [CrossRef] [PubMed]
- Kodaira, H.; Kusuhara, H.; Fujita, T.; Ushiki, J.; Fuse, E.; Sugiyama, Y. Quantitative Evaluation of the Impact of Active Efflux by P-Glycoprotein and Breast Cancer Resistance Protein at the Blood-Brain Barrier on the Predictability of the Unbound Concentrations of Drugs in the Brain Using Cerebrospinal Fluid Concentration as a Surrogate. J. Pharmacol. Exp. Ther. 2011, 339, 935–944. [Google Scholar] [CrossRef]
- Hasannejad, H.; Takeda, M.; Narikawa, S.; Huang, X.-L.; Enomoto, A.; Taki, K.; Niwa, T.; Jung, S.H.; Onozato, M.L.; Tojo, A.; et al. Human Organic Cation Transporter 3 Mediates the Transport of Antiarrhythmic Drugs. Eur. J. Pharmacol. 2004, 499, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Burckhardt, B.C.; Henjakovic, M.; Hagos, Y.; Burckhardt, G. Counter-Flow Suggests Transport of Dantrolene and 5-OH Dantrolene by the Organic Anion Transporters 2 (OAT2) and 3 (OAT3). Pflugers Arch. 2016, 468, 1909–1918. [Google Scholar] [CrossRef] [PubMed]
- Ronaldson, P.T.; Finch, J.D.; Demarco, K.M.; Quigley, C.E.; Davis, T.P. Inflammatory Pain Signals an Increase in Functional Expression of Organic Anion Transporting Polypeptide 1a4 at the Blood-Brain Barrier. J. Pharmacol. Exp. Ther. 2011, 336, 827–839. [Google Scholar] [CrossRef]
- Chen, X.; Loryan, I.; Payan, M.; Keep, R.F.; Smith, D.E.; Hammarlund-Udenaes, M. Effect of Transporter Inhibition on the Distribution of Cefadroxil in Rat Brain. Fluids Barriers CNS 2014, 11, 25. [Google Scholar] [CrossRef]
- Sekhar, G.N.; Georgian, A.R.; Sanderson, L.; Vizcay-Barrena, G.; Brown, R.C.; Muresan, P.; Fleck, R.A.; Thomas, S.A. Organic Cation Transporter 1 (OCT1) Is Involved in Pentamidine Transport at the Human and Mouse Blood-Brain Barrier (BBB). PLoS ONE 2017, 12, e0173474. [Google Scholar] [CrossRef]
- Mochizuki, T.; Mizuno, T.; Kurosawa, T.; Yamaguchi, T.; Higuchi, K.; Tega, Y.; Nozaki, Y.; Kawabata, K.; Deguchi, Y.; Kusuhara, H. Functional Investigation of Solute Carrier Family 35, Member F2, in Three Cellular Models of the Primate Blood-Brain Barrier. Drug Metab. Dispos. 2021, 49, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Reese, M.J.; Savina, P.M.; Generaux, G.T.; Tracey, H.; Humphreys, J.E.; Kanaoka, E.; Webster, L.O.; Harmon, K.A.; Clarke, J.D.; Polli, J.W. In Vitro Investigations into the Roles of Drug Transporters and Metabolizing Enzymes in the Disposition and Drug Interactions of Dolutegravir, a HIV Integrase Inhibitor. Drug Metab. Dispos. 2013, 41, 353–361. [Google Scholar] [CrossRef]
- EMA. EMA Assessment Report. Available online: https://www.ema.europa.eu/en/documents/assessment-report/dovato-epar-public-assessment-report_en.pdf (accessed on 4 October 2023).
- Janneh, O.; Chandler, B.; Hartkoorn, R.; Kwan, W.S.; Jenkinson, C.; Evans, S.; Back, D.J.; Owen, A.; Khoo, S.H. Intracellular Accumulation of Efavirenz and Nevirapine Is Independent of P-Glycoprotein Activity in Cultured CD4 T Cells and Primary Human Lymphocytes. J. Antimicrob. Chemother. 2009, 64, 1002–1007. [Google Scholar] [CrossRef]
- Hartkoorn, R.C.; Kwan, W.S.; Shallcross, V.; Chaikan, A.; Liptrott, N.; Egan, D.; Sora, E.S.; James, C.E.; Gibbons, S.; Bray, P.G.; et al. HIV Protease Inhibitors Are Substrates for OATP1A2, OATP1B1 and OATP1B3 and Lopinavir Plasma Concentrations Are Influenced by SLCO1B1 Polymorphisms. Pharmacogenet. Genom. 2010, 20, 112–120. [Google Scholar] [CrossRef]
- Jung, N.; Lehmann, C.; Rubbert, A.; Knispel, M.; Hartmann, P.; van Lunzen, J.; Stellbrink, H.-J.; Faetkenheuer, G.; Taubert, D. Relevance of the Organic Cation Transporters 1 and 2 for Antiretroviral Drug Therapy in Human Immunodeficiency Virus Infection. Drug Metab. Dispos. 2008, 36, 1616–1623. [Google Scholar] [CrossRef] [PubMed]
- Curley, P.; Rajoli, R.K.R.; Moss, D.M.; Liptrott, N.J.; Letendre, S.; Owen, A.; Siccardi, M. Efavirenz Is Predicted To Accumulate in Brain Tissue: An In Silico, In Vitro, and In Vivo Investigation. Antimicrob. Agents Chemother. 2017, 61, e01841-16. [Google Scholar] [CrossRef] [PubMed]
- Sato, S.; Matsumiya, K.; Tohyama, K.; Kosugi, Y. Translational CNS Steady-State Drug Disposition Model in Rats, Monkeys, and Humans for Quantitative Prediction of Brain-to-Plasma and Cerebrospinal Fluid-to-Plasma Unbound Concentration Ratios. AAPS J. 2021, 23, 81. [Google Scholar] [CrossRef]
- Khurana, V.; Minocha, M.; Pal, D.; Mitra, A.K. Role of OATP-1B1 and/or OATP-1B3 in Hepatic Disposition of Tyrosine Kinase Inhibitors. Drug Metab. Drug Interact. 2014, 29, 179–190. [Google Scholar] [CrossRef]
- Elmeliegy, M.A.; Carcaboso, A.M.; Tagen, M.; Bai, F.; Stewart, C.F. Role of ATP-Binding Cassette and Solute Carrier Transporters in Erlotinib CNS Penetration and Intracellular Accumulation. Clin. Cancer Res. 2011, 17, 89–99. [Google Scholar] [CrossRef]
- Bauer, M.; Karch, R.; Wulkersdorfer, B.; Philippe, C.; Nics, L.; Klebermass, E.-M.; Weber, M.; Poschner, S.; Haslacher, H.; Jäger, W.; et al. A Proof-of-Concept Study to Inhibit ABCG2- and ABCB1-Mediated Efflux Transport at the Human Blood–Brain Barrier. J. Nucl. Med. 2019, 60, 486–491. [Google Scholar] [CrossRef] [PubMed]
- Sampson, K.E.; Brinker, A.; Pratt, J.; Venkatraman, N.; Xiao, Y.; Blasberg, J.; Steiner, T.; Bourner, M.; Thompson, D.C. Zinc Finger Nuclease–Mediated Gene Knockout Results in Loss of Transport Activity for P-Glycoprotein, BCRP, and MRP2 in Caco-2 Cells. Drug Metab. Dispos. 2015, 43, 199–207. [Google Scholar] [CrossRef]
- Matsushima, S.; Maeda, K.; Hayashi, H.; Debori, Y.; Schinkel, A.H.; Schuetz, J.D.; Kusuhara, H.; Sugiyama, Y. Involvement of Multiple Efflux Transporters in Hepatic Disposition of Fexofenadine. Mol. Pharmacol. 2008, 73, 1474–1483. [Google Scholar] [CrossRef]
- Izumi, S.; Nozaki, Y.; Maeda, K.; Komori, T.; Takenaka, O.; Kusuhara, H.; Sugiyama, Y. Investigation of the Impact of Substrate Selection on In Vitro Organic Anion Transporting Polypeptide 1B1 Inhibition Profiles for the Prediction of Drug-Drug Interactions. Drug Metab. Dispos. 2015, 43, 235–247. [Google Scholar] [CrossRef]
- Shimizu, M.; Fuse, K.; Okudaira, K.; Nishigaki, R.; Maeda, K.; Kusuhara, H.; Sugiyama, Y. Contribution of OATP (organic anion-transporting polypeptide) family transporters to the hepatic uptake of fexofenadine in humans. Drug Metab. Dispos. 2005, 33, 1477–1481. [Google Scholar] [CrossRef]
- Ming, X.; Knight, B.M.; Thakker, D.R. Vectorial Transport of Fexofenadine across Caco-2 Cells: Involvement of Apical Uptake and Basolateral Efflux Transporters. Mol. Pharm. 2011, 8, 1677–1686. [Google Scholar] [CrossRef]
- Morita, T.; Akiyoshi, T.; Sato, R.; Uekusa, Y.; Katayama, K.; Yajima, K.; Imaoka, A.; Sugimoto, Y.; Kiuchi, F.; Ohtani, H. Citrus Fruit-Derived Flavanone Glycoside Narirutin Is a Novel Potent Inhibitor of Organic Anion-Transporting Polypeptides. J. Agric. Food Chem. 2020, 68, 14182–14191. [Google Scholar] [CrossRef] [PubMed]
- Glaeser, H.; Bailey, D.G.; Dresser, G.K.; Gregor, J.C.; Schwarz, U.I.; McGrath, J.S.; Jolicoeur, E.; Lee, W.; Leake, B.F.; Tirona, R.G.; et al. Intestinal Drug Transporter Expression and the Impact of Grapefruit Juice in Humans. Clin. Pharmacol. Ther. 2007, 81, 362–370. [Google Scholar] [CrossRef]
- Tahara, H.; Kusuhara, H.; Maeda, K.; Koepsell, H.; Fuse, E.; Sugiyama, Y. Inhibition of OAT3-Mediated Renal Uptake as a Mechanism for Drug-Drug Interaction Between Fexofenadine and Probenecid. Drug Metab. Dispos. 2006, 34, 743–747. [Google Scholar] [CrossRef] [PubMed]
- Mathialagan, S.; Piotrowski, M.A.; Tess, D.A.; Feng, B.; Litchfiled, J.; Varma, M.V. Quantitative Prediction of Human Renal Clearance and Drug-Drug Interactions of Organic Anion Transporter Substrates Using In Vitro Transport Data. Drug Metab. Dispos. 2017, 45, 409–417. [Google Scholar] [CrossRef]
- Matsushima, S.; Maeda, K.; Inoue, K.; Ohta, K.; Yuasa, H.; Kondo, T.; Nakayama, H.; Horita, S.; Kusuhara, H.; Sugiyama, Y. The Inhibition of Human Multidrug and Toxin Extrusion 1 Is Involved in the Drug-Drug Interaction Caused by Cimetidine. Drug Metab. Dispos. 2009, 37, 555–559. [Google Scholar] [CrossRef] [PubMed]
- Doan, K.M.; Wring, S.A.; Shampine, L.J.; Jordan, K.H.; Bishop, J.P.; Kratz, J.; Yang, E.; Serabjit-Singh, C.J.; Adkison, K.K.; Polli, J.W. Steady-State Brain Concentrations of Antihistamines in Rats. Pharmacology 2004, 72, 92–98. [Google Scholar] [CrossRef]
- Crowe, A.; Teoh, Y.-K. Limited P-Glycoprotein Mediated Efflux for Anti-Epileptic Drugs. J. Drug Target. 2006, 14, 291–300. [Google Scholar] [CrossRef]
- Cheng, Z.; Liu, H.; Yu, N.; Wang, F.; An, G.; Xu, Y.; Liu, Q.; Guan, C.; Ayrton, A. Hydrophilic Anti-Migraine Triptans Are Substrates for OATP1A2, a Transporter Expressed at Human Blood-Brain Barrier. Xenobiotica 2012, 42, 880–890. [Google Scholar] [CrossRef] [PubMed]
- Dickens, D.; Webb, S.D.; Antonyuk, S.; Giannoudis, A.; Owen, A.; Rädisch, S.; Hasnain, S.S.; Pirmohamed, M. Transport of Gabapentin by LAT1 (SLC7A5). Biochem. Pharmacol. 2013, 85, 1672–1683. [Google Scholar] [CrossRef] [PubMed]
- Feng, B.; Hurst, S.; Lu, Y.; Varma, M.V.; Rotter, C.J.; El-Kattan, A.; Lockwood, P.; Corrigan, B. Quantitative Prediction of Renal Transporter-Mediated Clinical Drug–Drug Interactions. Mol. Pharm. 2013, 10, 4207–4215. [Google Scholar] [CrossRef]
- Gebauer, L.; Jensen, O.; Brockmöller, J.; Dücker, C. Substrates and Inhibitors of the Organic Cation Transporter 3 and Comparison with OCT1 and OCT2. J. Med. Chem. 2022, 65, 12403–12416. [Google Scholar] [CrossRef]
- Pochini, L.; Galluccio, M.; Scalise, M.; Console, L.; Indiveri, C. OCTN: A Small Transporter Subfamily with Great Relevance to Human Pathophysiology, Drug Discovery, and Diagnostics. SLAS Discov. 2019, 24, 89–110. [Google Scholar] [CrossRef] [PubMed]
- Fridén, M.; Winiwarter, S.; Jerndal, G.; Bengtsson, O.; Wan, H.; Bredberg, U.; Hammarlund-Udenaes, M.; Antonsson, M. Structure-Brain Exposure Relationships in Rat and Human Using a Novel Data Set of Unbound Drug Concentrations in Brain Interstitial and Cerebrospinal Fluids. J. Med. Chem. 2009, 52, 6233–6243. [Google Scholar] [CrossRef]
- Redeker, K.-E.M.; Jensen, O.; Gebauer, L.; Meyer-Tönnies, M.J.; Brockmöller, J. Atypical Substrates of the Organic Cation Transporter 1. Biomolecules 2022, 12, 1664. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, N.; Furugen, A.; Ono, K.; Koishikawa, M.; Miyazawa, Y.; Nishimura, A.; Umazume, T.; Narumi, K.; Kobayashi, M.; Iseki, K. Cellular Uptake Properties of Lamotrigine in Human Placental Cell Lines: Investigation of Involvement of Organic Cation Transporters (SLC22A1–5). Drug Metab. Pharmacokinet. 2020, 35, 266–273. [Google Scholar] [CrossRef] [PubMed]
- Summerfield, S.G.; Lucas, A.J.; Porter, R.A.; Jeffrey, P.; Gunn, R.N.; Read, K.R.; Stevens, A.J.; Metcalf, A.C.; Osuna, M.C.; Kilford, P.J.; et al. Toward an Improved Prediction of Human In Vivo Brain Penetration. Xenobiotica 2008, 38, 1518–1535. [Google Scholar] [CrossRef]
- Uchida, Y.; Ohtsuki, S.; Kamiie, J.; Terasaki, T. Blood-Brain Barrier (BBB) Pharmacoproteomics: Reconstruction of In Vivo Brain Distribution of 11 P-Glycoprotein Substrates Based on the BBB Transporter Protein Concentration, In Vitro Intrinsic Transport Activity, and Unbound Fraction in Plasma and Brain in Mice. J. Pharmacol. Exp. Ther. 2011, 339, 579–588. [Google Scholar] [CrossRef]
- Jia, Y.; Liu, Z.; Wang, C.; Meng, Q.; Huo, X.; Liu, Q.; Sun, H.; Sun, P.; Yang, X.; Ma, X.; et al. P-Gp, MRP2 and OAT1/OAT3 Mediate the Drug-Drug Interaction between Resveratrol and Methotrexate. Toxicol. Appl. Pharmacol. 2016, 306, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Nozaki, Y.; Kusuhara, H.; Kondo, T.; Iwaki, M.; Shiroyanagi, Y.; Nakayama, H.; Horita, S.; Nakazawa, H.; Okano, T.; Sugiyama, Y. Species Difference in the Inhibitory Effect of Nonsteroidal Anti-Inflammatory Drugs on the Uptake of Methotrexate by Human Kidney Slices. J. Pharmacol. Exp. Ther. 2007, 322, 1162–1170. [Google Scholar] [CrossRef]
- Ramsey, L.B.; Bruun, G.H.; Yang, W.; Treviño, L.R.; Vattathil, S.; Scheet, P.; Cheng, C.; Rosner, G.L.; Giacomini, K.M.; Fan, Y.; et al. Rare versus Common Variants in Pharmacogenetics: SLCO1B1 Variation and Methotrexate Disposition. Genome Res. 2012, 22, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Abe, T.; Unno, M.; Onogawa, T.; Tokui, T.; Kondo, T.N.; Nakagomi, R.; Adachi, H.; Fujiwara, K.; Okabe, M.; Suzuki, T.; et al. LST-2, A Human Liver-Specific Organic Anion Transporter, Determines Methotrexate Sensitivity in Gastrointestinal Cancers. Gastroenterology 2001, 120, 1689–1699. [Google Scholar] [CrossRef] [PubMed]
- Visentin, M.; Chang, M.-H.; Romero, M.F.; Zhao, R.; Goldman, I.D. Substrate- and PH-Specific Antifolate Transport Mediated by Organic Anion-Transporting Polypeptide 2B1 (OATP2B1-SLCO2B1). Mol. Pharmacol. 2012, 81, 134–142. [Google Scholar] [CrossRef] [PubMed]
- Badagnani, I.; Castro, R.A.; Taylor, T.R.; Brett, C.M.; Huang, C.C.; Stryke, D.; Kawamoto, M.; Johns, S.J.; Ferrin, T.E.; Carlson, E.J.; et al. Interaction of Methotrexate with Organic-Anion Transporting Polypeptide 1A2 and Its Genetic Variants. J. Pharmacol. Exp. Ther. 2006, 318, 521–529. [Google Scholar] [CrossRef]
- Yuan, Y.; Yang, H.; Kong, L.; Li, Y.; Li, P.; Zhang, H.; Ruan, J. Interaction between Rhein Acyl Glucuronide and Methotrexate Based on Human Organic Anion Transporters. Chem.-Biol. Interact. 2017, 277, 79–84. [Google Scholar] [CrossRef]
- Kurata, T.; Iwamoto, T.; Kawahara, Y.; Okuda, M. Characteristics of Pemetrexed Transport by Renal Basolateral Organic Anion Transporter HOAT3. Drug Metab. Pharmacokinet. 2014, 29, 148–153. [Google Scholar] [CrossRef]
- Fujino, H.; Nakai, D.; Nakagomi, R.; Saito, M.; Tokui, T.; Kojima, J. Metabolic Stability and Uptake by Human Hepatocytes of Pitavastatin, a New Inhibitor of HMG-CoA Reductase. Arzneimittelforschung 2004, 54, 382–388. [Google Scholar] [CrossRef]
- Bi, Y.; Costales, C.; Mathialagan, S.; West, M.; Eatemadpour, S.; Lazzaro, S.; Tylaska, L.; Scialis, R.; Zhang, H.; Umland, J.; et al. Quantitative Contribution of Six Major Transporters to the Hepatic Uptake of Drugs: “SLC-Phenotyping” Using Primary Human Hepatocytes. J. Pharmacol. Exp. Ther. 2019, 370, 72–83. [Google Scholar] [CrossRef]
- Varma, M.V.; Rotter, C.J.; Chupka, J.; Whalen, K.M.; Duignan, D.B.; Feng, B.; Litchfield, J.; Goosen, T.C.; El-Kattan, A.F. PH-Sensitive Interaction of HMG-CoA Reductase Inhibitors (Statins) with Organic Anion Transporting Polypeptide 2B1. Mol. Pharm. 2011, 8, 1303–1313. [Google Scholar] [CrossRef]
- SHIRASAKA, Y.; SUZUKI, K.; SHICHIRI, M.; NAKANISHI, T.; TAMAI, I. Intestinal Absorption of HMG-CoA Reductase Inhibitor Pitavastatin Mediated by Organic Anion Transporting Polypeptide and P-Glycoprotein/Multidrug Resistance 1. Drug Metab. Pharmacokinet. 2011, 26, 171–179. [Google Scholar] [CrossRef]
- Yabuuchi, H.; Tamai, I.; Nezu, J.; Sakamoto, K.; Oku, A.; Shimane, M.; Sai, Y.; Tsuji, A. Novel Membrane Transporter OCTN1 Mediates Multispecific, Bidirectional, and PH-Dependent Transport of Organic Cations. J. Pharmacol. Exp. Ther. 1999, 289, 768–773. [Google Scholar] [PubMed]
- Grigat, S.; Fork, C.; Bach, M.; Golz, S.; Geerts, A.; Schömig, E.; Gründemann, D. The Carnitine Transporter SLC22A5 Is Not a General Drug Transporter, but It Efficiently Translocates Mildronate. Drug Metab. Dispos. 2009, 37, 330–337. [Google Scholar] [CrossRef] [PubMed]
- Tanihara, Y.; Masuda, S.; Sato, T.; Katsura, T.; Ogawa, O.; Inui, K. Substrate Specificity of MATE1 and MATE2-K, Human Multidrug and Toxin Extrusions/H+-Organic Cation Antiporters. Biochem. Pharmacol. 2007, 74, 359–371. [Google Scholar] [CrossRef] [PubMed]
- Hashiguchi, Y.; Hamada, A.; Shinohara, T.; Tsuchiya, K.; Jono, H.; Saito, H. Role of P-Glycoprotein in the Efflux of Raltegravir from Human Intestinal Cells and CD4+ T-Cells as an Interaction Target for Anti-HIV Agents. Biochem. Biophys. Res. Commun. 2013, 439, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Moss, D.M.; Kwan, W.S.; Liptrott, N.J.; Smith, D.L.; Siccardi, M.; Khoo, S.H.; Back, D.J.; Owen, A. Raltegravir Is a Substrate for SLC22A6: A Putative Mechanism for the Interaction between Raltegravir and Tenofovir. Antimicrob. Agents Chemother. 2011, 55, 879–887. [Google Scholar] [CrossRef] [PubMed]
- Srinivas, N.; Rosen, E.P.; Gilliland, W.M., Jr.; Kovarova, M.; Remling-Mulder, L.; Cruz, G.D.L.; White, N.; Adamson, L.; Schauer, A.P.; Sykes, C.; et al. Antiretroviral Concentrations and Surrogate Measures of Efficacy in the Brain Tissue and CSF of Preclinical Species. Xenobiotica 2019, 49, 1192–1201. [Google Scholar] [CrossRef]
- Collett, A.; Tanianis-Hughes, J.; Hallifax, D.; Warhurst, G. Predicting P-Glycoprotein Effects on Oral Absorption: Correlation of Transport in Caco-2 with Drug Pharmacokinetics in Wild-Type and Mdr1a(-/-) Mice In Vivo. Pharm. Res. 2004, 21, 819–826. [Google Scholar] [CrossRef]
- Wegler, C.; Gazit, M.; Issa, K.; Subramaniam, S.; Artursson, P.; Karlgren, M. Expanding the Efflux In Vitro Assay Toolbox: A CRISPR-Cas9 Edited MDCK Cell Line with Human BCRP and Completely Lacking Canine MDR1. J. Pharm. Sci. 2021, 110, 388–396. [Google Scholar] [CrossRef]
- Vavricka, S.R.; Montfoort, J.V.; Ha, H.R.; Meier, P.J.; Fattinger, K. Interactions of Rifamycin SV and Rifampicin with Organic Anion Uptake Systems of Human Liver. Hepatology 2002, 36, 164–172. [Google Scholar] [CrossRef] [PubMed]
- Te Brake, L.H.M.; van den Heuvel, J.J.M.W.; Buaben, A.O.; van Crevel, R.; Bilos, A.; Russel, F.G.; Aarnoutse, R.E.; Koenderink, J.B. Moxifloxacin Is a Potent In Vitro Inhibitor of OCT- and MATE-Mediated Transport of Metformin and Ethambutol. Antimicrob. Agents Chemother. 2016, 60, 7105–7114. [Google Scholar] [CrossRef] [PubMed]
- Deng, F.; Tuomi, S.-K.; Neuvonen, M.; Hirvensalo, P.; Kulju, S.; Wenzel, C.; Oswald, S.; Filppula, A.M.; Niemi, M. Comparative Hepatic and Intestinal Efflux Transport of Statins. Drug Metab. Dispos. 2021, 49, 750–759. [Google Scholar] [CrossRef]
- Wen, J.; Wei, X.; Sheng, X.; Zhou, D.; Peng, H.; Lu, Y.; Zhou, J. Effect of Ursolic Acid on Breast Cancer Resistance Protein-Mediated Transport of Rosuvastatin In Vivo and Vitro. Chin. Med. Sci. J. 2015, 30, 218–225. [Google Scholar] [CrossRef] [PubMed]
- Kimoto, E.; Li, R.; Scialis, R.J.; Lai, Y.; Varma, M.V.S. Hepatic Disposition of Gemfibrozil and Its Major Metabolite Gemfibrozil 1-O-β-Glucuronide. Mol. Pharm. 2015, 12, 3943–3952. [Google Scholar] [CrossRef]
- Bednarczyk, D.; Sanghvi, M.V. Organic Anion Transporting Polypeptide 2B1 (OATP2B1), an Expanded Substrate Profile, Does It Align with OATP2B1’s Hypothesized Function? Xenobiotica 2020, 50, 1128–1137. [Google Scholar] [CrossRef]
- Ronaldson, P.T.; Brzica, H.; Abdullahi, W.; Reilly, B.G.; Davis, T.P. Transport Properties of Statins by OATP1A2 and Regulation by Transforming Growth Factor-β (TGF-β) Signaling in Human Endothelial Cells. J. Pharmacol. Exp. Ther. 2020, 376. [Google Scholar] [CrossRef]
- Harati, R.; Benech, H.; Villégier, A.S.; Mabondzo, A. P-Glycoprotein, Breast Cancer Resistance Protein, Organic Anion Transporter 3, and Transporting Peptide 1a4 during Blood-Brain Barrier Maturation: Involvement of Wnt/β-Catenin and Endothelin-1 Signaling. Mol. Pharm. 2013, 10, 1566–1580. [Google Scholar] [CrossRef]
- Takeda, M.; Khamdang, S.; Narikawa, S.; Kimura, H.; Kobayashi, Y.; Yamamoto, T.; Cha, S.H.; Sekine, T.; Endou, H. Human Organic Anion Transporters and Human Organic Cation Transporters Mediate Renal Antiviral Transport. J. Pharmacol. Exp. Ther. 2002, 300, 918–924. [Google Scholar] [CrossRef]
- Minuesa, G.; Volk, C.; Molina-Arcas, M.; Gorboulev, V.; Erkizia, I.; Arndt, P.; Clotet, B.; Pastor-Anglada, M.; Koepsell, H.; Martinez-Picado, J. Transport of Lamivudine [(-)-β-l-2′,3′-Dideoxy-3′-Thiacytidine] and High-Affinity Interaction of Nucleoside Reverse Transcriptase Inhibitors with Human Organic Cation Transporters 1, 2, and 3. J. Pharmacol. Exp. Ther. 2009, 329, 252–261. [Google Scholar] [CrossRef] [PubMed]
- Parvez, M.M.; Kaisar, N.; Shin, H.J.; Jung, J.A.; Shin, J.-G. Inhibitory Interaction Potential of 22 Antituberculosis Drugs on Organic Anion and Cation Transporters of the SLC22A Family. Antimicrob. Agents Chemother. 2016, 60, 6558–6567. [Google Scholar] [CrossRef]
- Bors, L.A.; Erdő, F. Overcoming the Blood–Brain Barrier. Challenges and Tricks for CNS Drug Delivery. Sci. Pharm. 2019, 87, 6. [Google Scholar] [CrossRef]
- Singh, V.K.; Subudhi, B.B. Development and Characterization of Lysine-Methotrexate Conjugate for Enhanced Brain Delivery. Drug Deliv. 2016, 23, 2327–2337. [Google Scholar] [CrossRef] [PubMed]



| Transporter | Gene Name | Method | Protein Expression Level | Unit | Tissue/Cell | Reference | |
|---|---|---|---|---|---|---|---|
| Value | SD | ||||||
| OCT1 | SLC22A1 | Targeted LC-MS/MS | ULQ < 0.289 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] | |
| OCT1 | SLC22A1 | Targeted LC-MS/MS | ULQ < 0.288 | fmol/μg protein | Brain microvessels | [8] | |
| OCT1 | SLC22A1 | Targeted LC-MS/MS | 0.58 | 0.11 | pmol/mg protein | Brain microvessels | [6] |
| OCT1 | SLC22A1 | Targeted LC-MS/MS | 0.54 | 0.06 | pmol/mg protein | Brain microvessels | [6] |
| OCT2 | SLC22A2 | Targeted LC-MS/MS | ULQ < 0.254 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] | |
| OCT2 | SLC22A2 | Targeted LC-MS/MS | ULQ < 0.123 | fmol/μg protein | Brain microvessels | [8] | |
| OCT3 | SLC22A3 | Targeted LC-MS/MS | ULQ < 0.534 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] | |
| OCT3 | SLC22A3 | Targeted LC-MS/MS | ULQ < 0.207 | fmol/μg protein | Brain microvessels | [8] | |
| OCT3 | SLC22A3 | Targeted LC-MS/MS | 0.62 | 0.08 | pmol/mg protein | Brain microvessels | [6] |
| 4F2hc | SLC3A2 | Targeted LC-MS/MS | 1.42 | 0.28 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] |
| 4F2hc | SLC3A2 | Targeted LC-MS/MS | 3.47 | 0.83 | fmol/μg protein | Brain microvessels | [8] |
| 4F2hc | SLC3A2 | Targeted LC-MS/MS | 1.9 | 0.23 | fmol/μg protein | hCMEC/D3 (plasma membrane fraction) | [7] |
| ASBT | SLC10A2 | Targeted LC-MS/MS | ULQ < 0.288 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] | |
| ASBT | SLC10A2 | Targeted LC-MS/MS | ULQ < 0.12 | fmol/μg protein | Brain microvessels | [8] | |
| ASCT1 | SLC1A4 | Targeted LC-MS/MS | ULQ < 0.331 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] | |
| ASCT2 | SLC1A5 | Targeted LC-MS/MS | ULQ < 1.38 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] | |
| ASCT2 | SLC1A5 | Targeted LC-MS/MS | ULQ < 0.142 | fmol/μg protein | Brain microvessels | [8] | |
| ATA1 | SLC38A1 | Targeted LC-MS/MS | ULQ < 1.28 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] | |
| ATA1 | SLC38A1 | Targeted LC-MS/MS | ULQ < 0.175 | fmol/μg protein | Brain microvessels | [8] | |
| ATA1 | SLC38A1 | Targeted LC-MS/MS | 1.57 | 0.06 | fmol/μg protein | hCMEC/D3 (plasma membrane fraction) | [7] |
| ATA2 | SLC38A2 | Targeted LC-MS/MS | ULQ < 0.497 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] | |
| ATA2 | SLC38A2 | Targeted LC-MS/MS | ULQ < 0.143 | fmol/μg protein | Brain microvessels | [8] | |
| ATA3 | SLC38A4 | Targeted LC-MS/MS | ULQ < 0.823 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] | |
| ATA3 | SLC38A4 | Targeted LC-MS/MS | ULQ < 0.0656 | fmol/μg protein | Brain microvessels | [8] | |
| BGT1 | SLC6A12 | Targeted LC-MS/MS | ULQ < 1.95 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] | |
| BGT1 | SLC6A12 | Targeted LC-MS/MS | 3.16 | 0.94 | fmol/μg protein | Brain microvessels | [8] |
| BOCT | SLC22A17 | Targeted LC-MS/MS | ULQ < 0.265 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] | |
| BOIT | SLC22A17 | Targeted LC-MS/MS | ULQ < 0.503 | fmol/μg protein | Brain microvessels | [8] | |
| CAT1 | SLC7A1 | Targeted LC-MS/MS | 1.13 | 0.18 | fmol/μg protein | Brain microvessels | [8] |
| CAT1 | SLC7A1 | Targeted LC-MS/MS | 1.22 | 0.15 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] |
| CNT1 | SLC28A2 | Targeted LC-MS/MS | ULQ < 0.297 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] | |
| CNT1 | SLC28A1 | Targeted LC-MS/MS | ULQ < 0.308 | fmol/μg protein | Brain microvessels | [8] | |
| CNT2 | SLC28A2 | Targeted LC-MS/MS | ULQ < 0.867 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] | |
| CNT2 | SLC28A2 | Targeted LC-MS/MS | ULQ < 0.141 | fmol/μg protein | Brain microvessels | [8] | |
| CNT3 | SLC6A8 | Targeted LC-MS/MS | ULQ < 0.35 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] | |
| CNT3 | SLC28A3 | Targeted LC-MS/MS | ULQ < 0.552 | fmol/μg protein | Brain microvessels | [8] | |
| CRT1 | SLC6A8 | Targeted LC-MS/MS | ULQ < 0.0915 | fmol/μg protein | Brain microvessels | [8] | |
| CT2 | SLC22A16 | Targeted LC-MS/MS | ULQ < 0.122 | fmol/μg protein | Brain microvessels | [8] | |
| CTL1 | SLC44A1 | Targeted LC-MS/MS | ULQ < 0.293 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] | |
| CTL2 | SLC44A2 | Targeted LC-MS/MS | ULQ < 0.383 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] | |
| EAAT1 | SLC1A3 | Targeted LC-MS/MS | 5.04 | 0.18 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] |
| EAAT3 | SLC1A1 | Targeted LC-MS/MS | ULQ < 0.256 | fmol/μg protein | Brain microvessels | [8] | |
| EEAT1 | SLC1A3 | Targeted LC-MS/MS | 25.4 | 12.5 | fmol/μg protein | Brain microvessels | [8] |
| ENT1 | SLC29A1 | Targeted LC-MS/MS | 0.568 | 0.134 | fmol/μg protein | Brain microvessels | [8] |
| ENT1 | SLC29A1 | Targeted LC-MS/MS | 5.94 | 0.35 | fmol/μg protein | hCMEC/D3 (Plasma membrane fraction) | [7] |
| ENT1 | SLC29A1 | Targeted LC-MS/MS | 0.27 | 0.1 | pmol/mg protein | Brain microvessels | [6] |
| ENT1 | SLC29A1 | Targeted LC-MS/MS | 2.49 | 0.12 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] |
| ENT2 | SLC29A2 | Targeted LC-MS/MS | ULQ < 1.49 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] | |
| ENT2 | SLC29A2 | Targeted LC-MS/MS | ULQ < 0.18 | fmol/μg protein | Brain microvessels | [8] | |
| FATP1 | SLC27A1 | Targeted LC-MS/MS | ULQ < 1.04 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] | |
| FATP2 | SLC27A2 | Targeted LC-MS/MS | ULQ < 0.199 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] | |
| FATP3 | SLC27A3 | Targeted LC-MS/MS | ULQ < 0.44 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] | |
| FLIPT1 | SLC22A15 | Targeted LC-MS/MS | ULQ < 0.245 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] | |
| FLIPT1 | SLC22A15 | Targeted LC-MS/MS | ULQ < 0.101 | fmol/μg protein | Brain microvessels | [8] | |
| GAT2 | SLC6A13 | Targeted LC-MS/MS | ULQ < 1.3 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] | |
| GAT2 | SLC6A13 | Targeted LC-MS/MS | ULQ < 0.374 | fmol/μg protein | Brain microvessels | [8] | |
| GLUT2 | SLC2A2 | Targeted LC-MS/MS | ULQ < 5.85 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] | |
| GLUT4 | SLC2A4 | Targeted LC-MS/MS | ULQ < 2.52 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] | |
| GLUT4 | SLC2A4 | Targeted LC-MS/MS | ULQ < 0.136 | fmol/μg protein | Brain microvessels | [8] | |
| LAT1 | SLC7A5 | Targeted LC-MS/MS | ULQ < 0.76 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] | |
| LAT1 | SLC7A5 | Targeted LC-MS/MS | 0.431 | 0.091 | fmol/μg protein | Brain microvessels | [8] |
| LAT1 | SLC7A5 | Targeted LC-MS/MS | 0.59 | 0.15 | pmol/mg protein | Brain microvessels | [6] |
| LAT1 | SLC7A5 | Targeted LC-MS/MS | 0.71 | 0.25 | pmol/mg protein | Brain microvessels | [6] |
| LAT2 | SLC7A6 | Targeted LC-MS/MS | ULQ < 0.059 | fmol/μg protein | Brain microvessels | [8] | |
| LAT2 | SLC7A6 | Targeted LC-MS/MS | ULQ < 2.08 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] | |
| MATE1 | SLC47A1 | Targeted LC-MS/MS | ULQ < 0.33 | fmol/μg protein | Brain microvessels | [8] | |
| MATE1 | SLC47A2 | Targeted LC-MS/MS | 8.61 | 0.63 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] |
| MATE2-k | SLC47A2 | Targeted LC-MS/MS | ULQ < 2.19 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] | |
| MATE2-K | SLC47A2 | Targeted LC-MS/MS | ULQ < 0.295 | fmol/μg protein | Brain microvessels | [8] | |
| MCT1 | SLC16A1 | Targeted LC-MS/MS | 2.27 | 0.85 | fmol/μg protein | Brain microvessels | [8] |
| MCT1 | SLC16A1 | Targeted LC-MS/MS | 1.87 | 0.22 | fmol/μg protein | hCMEC/D3 (Plasma membrane fraction) | [7] |
| MCT1 | SLC16A1 | Targeted LC-MS/MS | 5.37 | 3.73 | pmol/mg protein | Brain microvessels | [6] |
| MCT1 | SLC16A1 | Targeted LC-MS/MS | 3.47 | 0.26 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] |
| MCT10 | SLC16A10 | Targeted LC-MS/MS | ULQ < 2.6 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] | |
| MCT2 | SLC16A7 | Targeted LC-MS/MS | ULQ < 0.671 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] | |
| MCT2 | SLC16A7 | Targeted LC-MS/MS | ULQ < 0.277 | fmol/μg protein | Brain microvessels | [8] | |
| MCT3 | SLC16A3 | Targeted LC-MS/MS | ULQ < 0.921 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] | |
| MCT4 | SLC16A4 | Targeted LC-MS/MS | 0.382 | 0.078 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] |
| MCT5 | SLC16A5 | Targeted LC-MS/MS | 0.685 | 0.124 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] |
| MCT8 | SLC16A2 | Targeted LC-MS/MS | 1.65 | 0.16 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] |
| MRP4 | ABCC4 | Targeted LC-MS/MS | 0.818 | 0.14 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] |
| NET | SLC6A2 | Targeted LC-MS/MS | ULQ < 0.361 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] | |
| NET | SLC6A2 | Targeted LC-MS/MS | ULQ < 0.441 | fmol/μg protein | Brain microvessels | [8] | |
| NTCP | SLC10A1 | Targeted LC-MS/MS | ULQ < 0.771 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] | |
| NTCP | SLC10A1 | Targeted LC-MS/MS | ULQ < 0.454 | fmol/μg protein | Brain microvessels | [8] | |
| OAT1 | SLC22A6 | Targeted LC-MS/MS | ULQ < 0.687 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] | |
| OAT1 | SLC22A6 | Targeted LC-MS/MS | ULQ < 0.909 | fmol/μg protein | Brain microvessels | [8] | |
| OAT1 | SLC22A6 | Targeted LC-MS/MS | 0.48 | 0.11 | pmol/mg protein | Brain microvessels | [6] |
| OAT2 | SLC22A7 | Targeted LC-MS/MS | ULQ < 0.152 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] | |
| OAT2 | SLC22A7 | Targeted LC-MS/MS | ULQ < 0.153 | fmol/μg protein | Brain microvessels | [8] | |
| OAT2 | SLC22A7 | Targeted LC-MS/MS | 7.9 | 3.8 | pmol/mg protein | Brain microvessels | [6] |
| OAT3 | SLC22A8 | Targeted LC-MS/MS | 1.87 | 0.12 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] |
| OAT3 | SLC22A8 | Targeted LC-MS/MS | ULQ < 0.348 | fmol/μg protein | Brain microvessels | [8] | |
| OAT3 | SLC22A8 | Targeted LC-MS/MS | 0.27 | 0.03 | pmol/mg protein | Brain microvessels | [6] |
| OAT4 | SLC22A11 | Targeted LC-MS/MS | ULQ < 0.534 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] | |
| OAT4 | SLC22A11 | Targeted LC-MS/MS | ULQ < 0.243 | fmol/μg protein | Brain microvessels | [8] | |
| OAT5 | SLC22A10 | Targeted LC-MS/MS | ULQ < 3.27 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] | |
| OAT5 | SLC22A10 | Targeted LC-MS/MS | ULQ < 0.0898 | fmol/μg protein | Brain microvessels | [8] | |
| OAT7 | SLC22A9 | Targeted LC-MS/MS | 0.51 | 0.1 | pmol/mg protein | Brain microvessels | [6] |
| OATP1 | SLCO | Targeted LC-MS/MS | 0.54 | 0.1 | pmol/mg protein | Brain microvessels | [6] |
| OATP1A2 | SLCO1A2 | Targeted LC-MS/MS | ULQ < 0.452 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] | |
| OATP1B1 | SLCO1B1 | Targeted LC-MS/MS | ULQ < 0.303 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] | |
| OATP1B3 | SLCO1B3 | Targeted LC-MS/MS | ULQ < 0.619 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] | |
| OATP1C1 | SLCO1C1 | Targeted LC-MS/MS | ULQ < 0.156 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] | |
| OATP1C1 | SLCO1C1 | Targeted LC-MS/MS | 0.27 | 0.03 | pmol/mg protein | Brain microvessels | [6] |
| OATP2B1 | SLCO2B1 | Targeted LC-MS/MS | ULQ < 0.237 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] | |
| OATP2B1 | SLCO2B1 | Targeted LC-MS/MS | 0.4 | 0.04 | pmol/mg protein | Brain microvessels | [6] |
| OATP2B1 | SLCO2B1 | Targeted LC-MS/MS | 0.48 | 0.11 | pmol/mg protein | Brain microvessels | [6] |
| OATP3A1 | SLCO3A1 | Targeted LC-MS/MS | 0.641 | 12 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] |
| OATP4A1 | SLCO4A1 | Targeted LC-MS/MS | ULQ < 1.2 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] | |
| OATP4C1 | SLCO4C1 | Targeted LC-MS/MS | ULQ < 0.283 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] | |
| OATP5A1 | SLCO5A1 | Targeted LC-MS/MS | ULQ < 3.28 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] | |
| OATP6A1 | SLCO6A1 | Targeted LC-MS/MS | ULQ < 0.545 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] | |
| OATP8 | SLCO1B3 | Targeted LC-MS/MS | 0.46 | 0.15 | pmol/mg protein | Brain microvessels | [6] |
| OATP-8 | SLCO1B3 | Targeted LC-MS/MS | ULQ < 0.572 | fmol/μg protein | Brain microvessels | [8] | |
| OATP-A | SLCO1A2 | Targeted LC-MS/MS | ULQ < 0.695 | fmol/μg protein | Brain microvessels | [8] | |
| OATP-B | SLCO2B1 | Targeted LC-MS/MS | ULQ < 0.337 | fmol/μg protein | Brain microvessels | [8] | |
| OATP-C | SLCO1B1 | Targeted LC-MS/MS | ULQ < 0.35 | fmol/μg protein | Brain microvessels | [8] | |
| OATP-D | SLCO3A1 | Targeted LC-MS/MS | ULQ < 0.254 | fmol/μg protein | Brain microvessels | [8] | |
| OATP-E | SLCO4A1 | Targeted LC-MS/MS | ULQ < 0.758 | fmol/μg protein | Brain microvessels | [8] | |
| OATP-F | SLCO1C1 | Targeted LC-MS/MS | ULQ < 0.208 | fmol/μg protein | Brain microvessels | [8] | |
| OATP-H | SLCO4C1 | Targeted LC-MS/MS | ULQ < 0.21 | fmol/μg protein | Brain microvessels | [8] | |
| OATP-I | SLCO | Targeted LC-MS/MS | ULQ < 0.082 | fmol/μg protein | Brain microvessels | [8] | |
| OATP-J | SLCO5A1 | Targeted LC-MS/MS | ULQ < 0.061 | fmol/μg protein | Brain microvessels | [8] | |
| OCTL1 | SLC22A13 | Targeted LC-MS/MS | ULQ < 0.532 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] | |
| OCTL1 | SLC22A13 | Targeted LC-MS/MS | ULQ < 0.699 | fmol/μg protein | Brain microvessels | [8] | |
| OCTL2 | SLC22A14 | Targeted LC-MS/MS | ULQ < 0.698 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] | |
| OCTL2 | SLC22A14 | Targeted LC-MS/MS | ULQ < 0.527 | fmol/μg protein | Brain microvessels | [8] | |
| OCTN1 | SLC22A4 | Targeted LC-MS/MS | ULQ < 0.25 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] | |
| OCTN1 | SLC22A4 | Targeted LC-MS/MS | ULQ < 0.123 | fmol/μg protein | Brain microvessels | [8] | |
| OCTN1 | SLC22A4 | Targeted LC-MS/MS | ULQ < 0.04 | 0.01 | pmol/mg protein | Brain microvessels | [6] |
| OCTN2 | SLC22A5 | Targeted LC-MS/MS | ULQ < 0.907 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] | |
| OCTN2 | SLC22A5 | Targeted LC-MS/MS | ULQ < 0.288 | fmol/μg protein | Brain microvessels | [8] | |
| OST-α | SLC51A | Targeted LC-MS/MS | 0.45 | 0.13 | pmol/mg protein | Brain microvessels | [6] |
| PCFT | SLC46A1 | Targeted LC-MS/MS | ULQ < 0.419 | fmol/μg protein | Brain microvessels | [8] | |
| PCFT | SLC46A1 | Targeted LC-MS/MS | 1.78 | 0.17 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] |
| PEPT1 | SLC15A1 | Targeted LC-MS/MS | ULQ < 0.325 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] | |
| PEPT1 | SLC15A1 | Targeted LC-MS/MS | ULQ < 0.379 | fmol/μg protein | Brain microvessels | [8] | |
| PEPT2 | SLC15A2 | Targeted LC-MS/MS | ULQ < 0.216 | fmol/μg protein | Brain microvessels | [8] | |
| PEPT2 | SLC15A2 | Targeted LC-MS/MS | ULQ < 0.37 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] | |
| PGT | SLCO2A1 | Targeted LC-MS/MS | ULQ < 0.233 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] | |
| PGT | SLCO2A1 | Targeted LC-MS/MS | ULQ < 0.186 | fmol/μg protein | Brain microvessels | [8] | |
| PHT2 | SLC15A3 | Targeted LC-MS/MS | ULQ < 0.456 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] | |
| PMAT | SLC29A4 | Targeted LC-MS/MS | ULQ < 0.191 | fmol/μg protein | Brain microvessels | [8] | |
| PMAT | SLC29A4 | Targeted LC-MS/MS | 0.288 | 0.041 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] |
| RFC | SLC19A | Targeted LC-MS/MS | 0.76 | 0.04 | fmol/μg protein | Brain microvessels | [8] |
| RFC | SLC19A | Targeted LC-MS/MS | 0.76 | 0.04 | fmol/μg protein | Brain microvessels | [8] |
| RFC1 | SLC19A1 | Targeted LC-MS/MS | 3.68 | 0.09 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] |
| SERT | SLC6A4 | Targeted LC-MS/MS | ULQ < 0.304 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] | |
| SERT | SLC6A4 | Targeted LC-MS/MS | ULQ < 0.116 | fmol/μg protein | Brain microvessels | [8] | |
| SLC22A18 | SLC22A18 | Targeted LC-MS/MS | ULQ < 0.375 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] | |
| SLC22A18 | SLC22A18 | Targeted LC-MS/MS | ULQ < 0.345 | fmol/μg protein | Brain microvessels | [9] | |
| TAUT | SLC6A6 | Targeted LC-MS/MS | ULQ < 0.169 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] | |
| TAUT | SLC6A6 | Targeted LC-MS/MS | ULQ < 0.0767 | fmol/μg protein | Brain microvessels | [8] | |
| TfR1 | TFRC | Targeted LC-MS/MS | 2.34 | 0.76 | fmol/μg protein | Brain microvessels | [8] |
| URAT1 | SLC22A12 | Targeted LC-MS/MS | ULQ < 0.357 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] | |
| URAT1 | SLC22A12 | Targeted LC-MS/MS | ULQ < 0.0566 | fmol/μg protein | Brain microvessels | [8] | |
| UST3 | SLC22A9 | Targeted LC-MS/MS | ULQ < 1.21 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] | |
| UST3 | SLC22A9 | Targeted LC-MS/MS | ULQ < 0.326 | fmol/μg protein | Brain microvessels | [8] | |
| xCT | SLC7A11 | Targeted LC-MS/MS | ULQ < 0.783 | fmol/μg protein | Choroid plexus (Plasma membrane fraction) | [9] | |
| xCT | SLC7A11 | Targeted LC-MS/MS | ULQ < 0.429 | fmol/μg protein | Brain microvessels | [8] | |
| Species | Transporter | Substrate | Perpetrator | Method | Exposure | DDI/Effect | Expression Level (fmol/μg Protein) | Reference |
|---|---|---|---|---|---|---|---|---|
| Mouse | Oatp1a4 | Glyburide | Rifampicin | In situ brain perfusion | Kin (0.5 ± 0.11 μL/g/s) | No change (Kin: ~0.4) | [87] | |
| Mouse | Oatp1a4 | Rosuvastatin | - | In vivo brain uptake | 57 ± 9 μL/min/g brain (brain/plasma = 22 μL/g) | - | [88] | |
| Pravastatin | 24 ± 4 μL/min/g brain (brain/plasma = 8.3 μL/g) | |||||||
| Taurocholate | 11 ± 2 μL/min/g brain | |||||||
| Ochratoxin A | 11 ± 0 μL/min/g brain (brain/plasma = 7.6 μL/g) | |||||||
| Mouse (Bend.3) | OCT1, OCT2, OCT3 | Pentamidine | Amantadine (500 μM) | In vitro uptake (Vd) | Vd at different time points | 59% Reduction | [96] | |
| Prazosin (100 μM) | No change (paracellular leakage increased) | |||||||
| N-methy-l nicotinamide (100 μM) | No change | |||||||
| Rat | Oatp1a4 | - | - | - | - | 1.99 | [49] | |
| Rat | Oct1/2 | Memantine (5 mh/kg i.v.) | Cimetidine (25 μM) | In vivo brain uptake | 84.59 ± 9.73 pmol/mg brain tissue | 37% Decreased (54.14 ± 8.35) | - | [82] |
| Rat | Oatp1a4 | Atorvastatin | BMP-9 (1 μg/kg) | In vivo brain uptake | AUC: 987.9 ± 53.41 pmol × min/mg brain tissue | 60% Increased (1581 ± 52.26) | - | [83] |
| LDN (10 mg/kg) + BMP-9 | Attenuated the BMP-9 effect | |||||||
| Pravastatin | BMP-9 (1 μg/kg) | AUC: 800.0 ± 47.41 pmol × min/mg brain tissue | 69% Increased (1349.00 ± 48.00) | |||||
| LDN (10 mg/kg) + BMP-9 | Attenuated the BMP-9 effect | |||||||
| Rosuvastatin | BMP-9 (1 μg/kg) | AUC: 836.8 ± 50.53 pmol × min/mg brain tissue | 74% Increased (1459.0 ± 53.51) | |||||
| LDN (10 mg/kg) + BMP-9 | Attenuated the BMP-9 effect | |||||||
| Rat | Oatp1a4 | Atorvastatin | Fexofenadine (100 μM) | In situ brain perfusion | 63.72 ± 9.78 pmol/mg brain tissue | 39% reduced (24.89 ± 7.55) | - | |
| Pravastatin | Fexofenadine (100 μM) | 54.98 ± 6.37 pmol/mg brain tissue | Reduced (12.39 ± 4.8) | |||||
| Rosuvastatin | Fexofenadine (100 μM) | 55.83 ± 7.84 pmol/mg brain tissue | Reduce (10.54 ± 3.65) | |||||
| Rat | Octs | SHY-01 (50 mg/kg) | - | In vivo | 2.05 ± 0.18 (hr·μg/mL) | CL: 24.48 ± 2.25 | - | [84] |
| Metformin (50 mg/kg) | - | In vivo | 1.89 ± 0.08 (hr·μg/mL) | CL: 26.46 ± 1.10 | ||||
| Rat | Oatp | Digoxin (2 mg/kg, i.v.) | Rifampicin (30 mg/kg, oral) | In vivo | ~0.07 (Kp,AUC,brain) | Increased (~1.8-fold) | - | [85] |
| ~0.02 (Kp,AUC,CSF) | Increased (~4-fold) | |||||||
| Rat | Oatp1a4 | Taurocholate | BMP-9 (1 μg/kg) | In vivo brain uptake | AUC: 1143.6 ± 57.92 pmol × min/mg brain tissue) | 79% Increased (2054.83 ± 66.13) | - | [89] |
| E3S (100 μM) | In situ brain perfusion | 65.31 ± 8.19 pmol/mg | 59% Reduced (27.02 ± 7.56) | |||||
| Fexofenadine (100 μM) | 61% Reduced (25.61 ± 7.44) | |||||||
| BSP | No effect (66.81 ± 7.13) | |||||||
| Atorvastatin | E3S (100 μM) | 34.07 ± 5.67 pmol/mg brain tissue | Reduced (17.67 ± 5.22 pmol/mg brain tissue) | |||||
| Pravastatin | 22.01 ± 6.27 pmol/mg brain tissue | Reduced (9.00 ± 4.98 pmol/mg brain tissue) | ||||||
| Rat | Oatp1a4 | Taurocholate | E3S (100 μM) | In situ brain perfusion | ~55 pmol/g brain tissue | Reduced (2.2-fold) | - | [94] |
| Digoxin (200 μM) | Reduced (2.4-fold) | |||||||
| Fexofenadine (100 μM) | Vbrain = 97.61 ± pmol/g | Reduced (2.2-fold) | ||||||
| BSP | No effect | |||||||
| Rat | Oats, Mrps, Oatps | Cefadroxil | Probenecid | Microdialysis | AUCblood = 1802 ± 97 (μg × min/mL) | 2873 ± 177 (Increased) | - | [95,97] |
| AUCECF = 40 ± 7 | 174 ± 35 (Increased) | |||||||
| Kp,uu,ECF = 0.022 ± 0.003 | 0.058 ± 0.009 (Increased) | |||||||
| AUCCSF = 57 ± 15 | 117 ± 50 (Increased) | |||||||
| Kp,uu,CSF = 0.031 ± 0.007 | 0.039 ± 0.015 | |||||||
| Pept2 | Cefadroxil | Ala-Ala | Brain slices | V,u,brain (mL/g brain) = 3.67 ± 0.23 | 0.95 ± 0.45 (Reduced) | |||
| GlySar | 1.10 ± 0.05 mL/g (Reduced) | |||||||
| Oats, Mrps, Oatps | Probenecid | 6.06 ± 0.15 (Increased) | ||||||
| Dog | OCT2 | - | LC-MS/MS | - | - | <LOQ | [90,92,93] | |
| - | - | - | - | <LOQ | ||||
| OAT3 | - | - | - | - | <LOQ | |||
| - | - | - | - | <LOQ | ||||
| OATP1A2 | - | - | - | - | <LOQ | |||
| - | - | - | - | 2.69 ± 0.78 | ||||
| OATP2B1 | - | - | - | - | <LOQ | |||
| - | - | - | - | <LOQ | ||||
| ENT1 | - | - | - | - | 0.581 ± 0.342 | |||
| - | - | - | - | 1.05 ± 0.47 | ||||
| LAT1 | - | - | - | - | <LOQ | |||
| - | - | - | - | <LOQ | ||||
| OCT3/P-gp | Quinidine (7.71 μmol/kg) | - | In vivo brain uptake | K,p,uu,brain = 0.363 ± 0.11 | - | - | ||
| - | K,p,uu,csf = 0.131 ± 0.036 | - | ||||||
| OAT2/BCRP | Dantrolene (1.59 μmol/kg) | - | Kp,uu,brain = 0.0614 ± 0.0021 | - | - | |||
| - | K,p,uu,csf = 0.505 ± 0.025 | - | ||||||
| Monkey (Baboon) | OATP2B1, OATP1A2 | Glyburide | Rifampicin | PET | 4.5 ± 1.0 (AUCbrain/AUCblood = 0.032) | No change: 11.5 (0.018) | - | [87] |
| Cyclosporine | No change: 17.2 (0.029) | |||||||
| Pantoprazole | No change: 8.1 (0.035) | |||||||
| Monkey | OATP2B1 | - | - | - | - | 0.12 | [49] | |
| Human (hCMEC/D3) | OCT1, OCT2, OCT3 | Pentamidine | Amantadine (500 μM) | In vitro uptake (Vd) | Vd at different time points | 45% Reduction | - | [96] |
| Prazosin (100 μM) | 39% Reduction | |||||||
| N-methy-l nicotinamide (100 μM) | No change | |||||||
| Human | OATP2B1, OATP1A2 | Glyburide | Rifampicin (9 mg/kg i.v.) | PET | 5.82 ± 0.74 (AUCbrain/AUCblood = 0.03) | No change: 7.72 (0.03) | - | [86] |
| Drug Name | Drug Class | Mw a | LogD b | Plasma Protein Binding (%) a | Efflux Transporter Substrate | Uptake Transporter Substrate | Kp,uu,brain | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MDR1 | BCRP | OATP1B1 | OATP1B3 | OATP2B1 | OATP1A2 | OCT1 | OCT2 | OCT3 | OCTN1 | OCTN2 | LAT1 | OAT1 | OAT3 | MATE1 | MATE2k | Rat | Mouse | Monkey | Human | |||||
| Dolutegravir | HIV-Integrase strand transfer inhibitor | 419.38 | 1.10 | 98.90 | Yes [98] | Yes [98] | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 0.02 [99] | NA | NA | NA |
| Efavirenz | Non-nucleoside Reverse transcriptase inhibitors (NNRTI) | 315.68 | 4.46 | 99.60 | No [100] | NA | No [101] | No [101] | NA | No [101] | No [102] | No [102] | NA | NA | NA | NA | NA | NA | NA | NA | 0.20 [103] | NA | NA | NA |
| Erlotinib | Kinase inhibitor | 393.40 | 3.05 | 93.00 | Yes [104] | Yes [104] | No [105] | No [105] | Yes [54] | NA | No [106] | Yes [106] | NA | NA | NA | NA | No [106] | Yes [106] | NA | NA | 0.06 [104] | NA | 0.05 [104] | 0.08 [107] |
| Fexofenadine | H-1 Receptor antagonists | 501.66 | 2.93 | 65.00 | Yes [108] | No [109] | Yes [110] | Yes [111] | Yes [112] | Yes [113] | Yes [114] | No [115] | No [112] | NA | No [112] | NA | Yes [116] | Yes [116] | Yes [117] | Yes [117] | 0.05 [118] | 0.22 [26] | NA | NA |
| Gabapentin | Anticonvulsant | 171.20 | −1.27 | <3 | No [119] | NA | NA | NA | NA | No [120] | No [121] | Yes [122] | No [123] | Yes [124] | No [121] | Yes [122] | No [122] | No [122] | NA | NA | 0.14 [125] | NA | NA | 0.16 [125] |
| Lamotrigine | Anticonvulsants | 256.10 | 1.91 | 55.00 | No [104] | No [104] | NA | NA | NA | No [120] | Yes [126] | Yes [126] | Yes [126] | No [127] | No [127] | NA | NA | NA | NA | NA | 0.88 [125] | NA | 0.86 [104] | 2.80 [128] |
| Loperamide | Antidiarrheal | 477.10 | 2.77 | 95.00 | Yes [104] | No [104] | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 0.02 [128] | NA | 0.04 [129] | NA |
| Methotrexate | Antimetabolite | 454.40 | −6.56 | 50.25 | Yes [130] | Yes [131] | Yes [132] | Yes [133] | Yes [134] | Yes [134] | NA | Yes [135] | NA | NA | NA | NA | Yes [136] | Yes [137] | Yes [135] | Yes [135] | 0.006 [125] | NA | 0.04 [104] | NA |
| Pitavastatin | HMG CoA Reductase Inhibitors (statin) | 421.50 | 0.89 | >99 | Yes [104] | Yes [104] | Yes [138] | Yes [139] | Yes [140] | Yes [141] | No | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 0.24 [104] | NA |
| Quinidine | Antiarrhythmic | 324.40 | 0.86 | 78.00 | Yes [104] | No [104] | NA | NA | NA | NA | NA | NA | Yes [92] | Yes [142] | No [143] | NA | NA | NA | No [144] | No [144] | 0.04 [104] | NA | 0.10 [104] | NA |
| Raltegravir | HIV-Integrase Strand transfer inhibitor | 444.42 | −0.92 | 83.00 | Yes [5] | Yes [5] | No [145] | No [145] | NA | No [145] | No [145] | NA | NA | NA | No [145] | NA | Yes [146] | NA | NA | NA | 0.13 [147] | NA | 0.12 [147] | NA |
| Rifampicin | Antibiotic | 822.90 | 2.87 | 89.00 | Yes [148] | No [149] | Yes [150] | Yes [150] | No [150] | No [150] | No [102] | No [102] | No [151] | NA | NA | NA | NA | NA | NA | NA | 0.04 [125] | NA | NA | NA |
| Rosuvastatin | HMG CoA Reductase Inhibitors (statin) | 481.54 | −1.24 | 88.00 | Yes [152] | Yes [153] | Yes [154] | Yes [154] | Yes [155] | Yes [156] | No [139] | NA | NA | NA | NA | NA | NA | NA | NA | NA | 3.97 [157] | NA | NA | NA |
| Zidovudine | Nucleoside Reverse Transcriptase Inhibitors (NRTI) | 267.20 | −0.41 | <38 | Yes [104] | Yes [104] | NA | NA | NA | NA | No [158] | No [158] | No [159] | NA | NA | NA | Yes [160] | Yes [158] | NA | NA | 0.09 [125] | NA | NA | NA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parvez, M.M.; Sadighi, A.; Ahn, Y.; Keller, S.F.; Enoru, J.O. Uptake Transporters at the Blood–Brain Barrier and Their Role in Brain Drug Disposition. Pharmaceutics 2023, 15, 2473. https://doi.org/10.3390/pharmaceutics15102473
Parvez MM, Sadighi A, Ahn Y, Keller SF, Enoru JO. Uptake Transporters at the Blood–Brain Barrier and Their Role in Brain Drug Disposition. Pharmaceutics. 2023; 15(10):2473. https://doi.org/10.3390/pharmaceutics15102473
Chicago/Turabian StyleParvez, Md Masud, Armin Sadighi, Yeseul Ahn, Steve F. Keller, and Julius O. Enoru. 2023. "Uptake Transporters at the Blood–Brain Barrier and Their Role in Brain Drug Disposition" Pharmaceutics 15, no. 10: 2473. https://doi.org/10.3390/pharmaceutics15102473
APA StyleParvez, M. M., Sadighi, A., Ahn, Y., Keller, S. F., & Enoru, J. O. (2023). Uptake Transporters at the Blood–Brain Barrier and Their Role in Brain Drug Disposition. Pharmaceutics, 15(10), 2473. https://doi.org/10.3390/pharmaceutics15102473