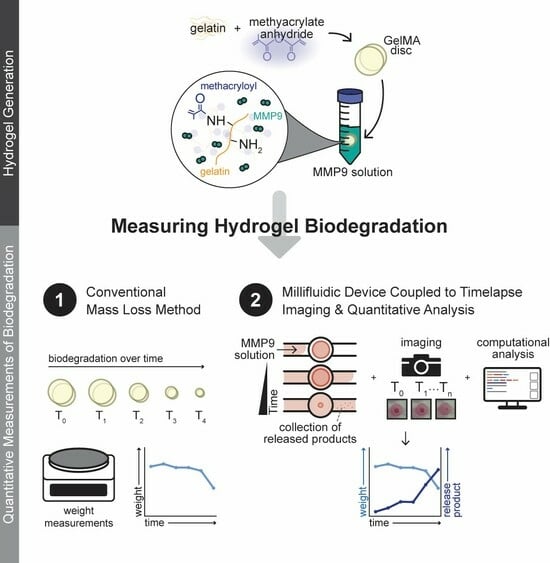
A Rapid Screening Platform for Simultaneous Evaluation of Biodegradation and Therapeutic Release of an Ocular Hydrogel
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of GelMA
2.2. Preparation of GelMA-PVA Hydrogels
2.3. Biodegradation of GelMA with MMP9 by Mass Loss in a Vial
2.4. Biodegradation of GelMA with MMP9 Using a Custom-Designed Millifluidic Device System
2.5. Image Analysis of Hydrogel Degradation
2.6. Estimation of Average Biodegradation Rates
2.7. Release of PVA from Millifluidic Chip
2.8. Detection of PVA
2.9. Quantification and Statistical Analysis
3. Results
3.1. Combined Millifluidics Imaging and Computational Analysis for Quantitative Characterization of GelMA Biodegradation
3.2. Release of Model Therapeutic PVA Can Be Quantified from the Millifluidic Device
4. Discussion
4.1. Novel Quantitative Millifluidic Imaging System to Quantify Hydrogel Biodegradation
4.2. Potential for High-Throughput Screening of Hydrogels Used for Drug Delivery
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lanier, O.L.; Manfre, M.G.; Bailey, C.; Liu, Z.; Sparks, Z.; Kulkarni, S.; Chauhan, A. Review of Approaches for Increasing Ophthalmic Bioavailability for Eye Drop Formulations. AAPS PharmSciTech 2021, 22, 107. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.; Amin, M.M.; Sayed, S. Ocular Drug Delivery: A Comprehensive Review. AAPS PharmSciTech 2023, 24, 66. [Google Scholar] [CrossRef] [PubMed]
- Mofidfar, M.; Abdi, B.; Ahadian, S.; Mostafavi, E.; Desai, T.A.; Abbasi, F.; Sun, Y.; Manche, E.E.; Ta, C.N.; Flowers, C.W. Drug Delivery to the Anterior Segment of the Eye: A Review of Current and Future Treatment Strategies. Int. J. Pharm. 2021, 607, 120924. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Li, S.; Xu, M.; Zhong, Y.; Fan, W.; Xu, J.; Zhou, T.; Ji, J.; Ye, J.; Yao, K. Polymer- and Lipid-Based Nanocarriers for Ocular Drug Delivery: Current Status and Future Perspectives. Adv. Drug Deliv. Rev. 2023, 196, 114770. [Google Scholar] [CrossRef]
- Suri, R.; Beg, S.; Kohli, K. Target Strategies for Drug Delivery Bypassing Ocular Barriers. J. Drug Deliv. Sci. Technol. 2020, 55, 101389. [Google Scholar] [CrossRef]
- Lynch, C.R.; Kondiah, P.P.D.; Choonara, Y.E.; du Toit, L.C.; Ally, N.; Pillay, V. Hydrogel Biomaterials for Application in Ocular Drug Delivery. Front. Bioeng. Biotechnol. 2020, 8, 228. [Google Scholar] [CrossRef]
- Yang, C.; Yang, J.; Lu, A.; Gong, J.; Yang, Y.; Lin, X.; Li, M.; Xu, H. Nanoparticles in Ocular Applications and Their Potential Toxicity. Front. Mol. Biosci. 2022, 9, 931759. [Google Scholar] [CrossRef]
- Jones, L.; Hui, A.; Phan, C.M.; Read, M.L.; Azar, D.; Buch, J.; Ciolino, J.B.; Naroo, S.A.; Pall, B.; Romond, K.; et al. CLEAR—Contact Lens Technologies of the Future. Cont. Lens Anterior Eye 2021, 44, 398–430. [Google Scholar] [CrossRef]
- Kamaly, N.; Yameen, B.; Wu, J.; Farokhzad, O.C. Degradable Controlled-Release Polymers and Polymeric Nanoparticles: Mechanisms of Controlling Drug Release. Chem. Rev. 2016, 116, 2602. [Google Scholar] [CrossRef]
- Senapati, S.; Mahanta, A.K.; Kumar, S.; Maiti, P. Controlled Drug Delivery Vehicles for Cancer Treatment and Their Performance. Signal Transduct. Target. Ther. 2018, 3, 7. [Google Scholar] [CrossRef]
- Petersen, R.S.; Nielsen, L.H.; Rindzevicius, T.; Boisen, A.; Keller, S.S. Controlled Drug Release from Biodegradable Polymer Matrix Loaded in Microcontainers Using Hot Punching. Pharmaceutics 2020, 12, 1050. [Google Scholar] [CrossRef] [PubMed]
- Benhabbour, S.R.; Kovarova, M.; Jones, C.; Copeland, D.J.; Shrivastava, R.; Swanson, M.D.; Sykes, C.; Ho, P.T.; Cottrell, M.L.; Sridharan, A.; et al. Ultra-Long-Acting Tunable Biodegradable and Removable Controlled Release Implants for Drug Delivery. Nat. Commun. 2019, 10, 4324. [Google Scholar] [CrossRef] [PubMed]
- Welling, C.; Schwengler, H.; Strahl, B. In-Vitro Degradation Test for Screening of Biomaterials. In Degradation Phenomena on Polymeric Biomaterials; Springer: Berlin/Heidelberg, Germany, 1992; pp. 25–36. [Google Scholar] [CrossRef]
- Hadavi, E.; de Vries, R.H.W.; Smink, A.M.; de Haan, B.; Leijten, J.; Schwab, L.W.; Karperien, M.H.B.J.; de Vos, P.; Dijkstra, P.J.; van Apeldoorn, A.A. In Vitro Degradation Profiles and In Vivo Biomaterial–Tissue Interactions of Microwell Array Delivery Devices. J. Biomed. Mater. Res. B Appl. Biomater. 2021, 109, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Azevedo, H.S.; Gama, F.M.; Reis, R.L. In Vitro Assessment of the Enzymatic Degradation of Several Starch Based Biomaterials. Biomacromolecules 2003, 4, 1703–1712. [Google Scholar] [CrossRef]
- Phan, C.M.; Subbaraman, L.N.; Jones, L. In Vitro Drug Release of Natamycin from β-Cyclodextrin and 2-Hydroxypropyl-β-Cyclodextrin-Functionalized Contact Lens Materials. J. Biomater. Sci. Polym. Ed. 2014, 25, 1907–1919. [Google Scholar] [CrossRef]
- Peng, C.C.; Kim, J.; Chauhan, A. Extended Delivery of Hydrophilic Drugs from Silicone-Hydrogel Contact Lenses Containing Vitamin E Diffusion Barriers. Biomaterials 2010, 31, 4032–4047. [Google Scholar] [CrossRef]
- Pereira-da-Mota, A.F.; Vivero-Lopez, M.; Garg, P.; Phan, C.M.; Concheiro, A.; Jones, L.; Alvarez-Lorenzo, C. In Vitro–In Vivo Correlation of Drug Release Profiles from Medicated Contact Lenses Using an In Vitro Eye Blink Model. Drug Deliv. Transl. Res. 2023, 13, 1116–1127. [Google Scholar] [CrossRef]
- Wu, Q.; Liu, J.; Wang, X.; Feng, L.; Wu, J.; Zhu, X.; Wen, W.; Gong, X. Organ-on-a-Chip: Recent Breakthroughs and Future Prospects. Biomed. Eng. Online 2020, 19, 9. [Google Scholar] [CrossRef]
- Leung, C.M.; de Haan, P.; Ronaldson-Bouchard, K.; Kim, G.A.; Ko, J.; Rho, H.S.; Chen, Z.; Habibovic, P.; Jeon, N.L.; Takayama, S.; et al. A Guide to the Organ-on-a-Chip. Nat. Rev. Methods Primers 2022, 2, 33. [Google Scholar] [CrossRef]
- Dittrich, P.S.; Manz, A. Lab-on-a-Chip: Microfluidics in Drug Discovery. Nat. Rev. Drug Discov. 2006, 5, 210–218. [Google Scholar] [CrossRef]
- Liu, Y.; Sun, L.; Zhang, H.; Shang, L.; Zhao, Y. Microfluidics for Drug Development: From Synthesis to Evaluation. Chem. Rev. 2021, 121, 7468–7529. [Google Scholar] [CrossRef] [PubMed]
- Staicu, C.E.; Jipa, F.; Axente, E.; Radu, M.; Radu, B.M.; Sima, F. Lab-on-a-Chip Platforms as Tools for Drug Screening in Neuropathologies Associated with Blood–Brain Barrier Alterations. Biomolecules 2021, 11, 916. [Google Scholar] [CrossRef] [PubMed]
- Maurya, R.; Gohil, N.; Bhattacharjee, G.; Alzahrani, K.J.; Ramakrishna, S.; Singh, V. Microfluidics Device for Drug Discovery, Screening and Delivery. Prog. Mol. Biol. Transl. Sci. 2022, 187, 335–346. [Google Scholar] [CrossRef] [PubMed]
- Ramasamy, M.; Ho, B.; Phan, C.M.; Qin, N.; Ren, C.L.; Jones, L. Inexpensive and Rapid Fabrication of PDMS Microfluidic Devices for Biological Testing Applications Using Low Cost Commercially Available 3D Printers. J. Micromechanics Microengineering 2023, 33, 105016. [Google Scholar] [CrossRef]
- Bhattacharjee, N.; Urrios, A.; Kang, S.; Folch, A. The Upcoming 3D-Printing Revolution in Microfluidics. Lab. Chip 2016, 16, 1720. [Google Scholar] [CrossRef]
- Bupphathong, S.; Quiroz, C.; Huang, W.; Chung, P.F.; Tao, H.Y.; Lin, C.H. Gelatin Methacrylate Hydrogel for Tissue Engineering Applications—A Review on Material Modifications. Pharmaceuticals 2022, 15, 171. [Google Scholar] [CrossRef]
- Piao, Y.; You, H.; Xu, T.; Bei, H.P.; Piwko, I.Z.; Kwan, Y.Y.; Zhao, X. Biomedical Applications of Gelatin Methacryloyl Hydrogels. Eng. Regen. 2021, 2, 47–56. [Google Scholar] [CrossRef]
- Akkurt Arslan, M.; Brignole-Baudouin, F.; Chardonnet, S.; Pionneau, C.; Blond, F.; Baudouin, C.; Kessal, K. Profiling Tear Film Enzymes Reveals Major Metabolic Pathways Involved in the Homeostasis of the Ocular Surface. Sci. Rep. 2023, 13, 15231. [Google Scholar] [CrossRef]
- Smith, V.A.; Rishmawi, H.; Hussein, H.; Easty, D.L. Tear Film MMP Accumulation and Corneal Disease. Br. J. Ophthalmol. 2001, 85, 147–153. [Google Scholar] [CrossRef]
- Teodorescu, M.; Bercea, M.; Morariu, S. Biomaterials of PVA and PVP in Medical and Pharmaceutical Applications: Perspectives and Challenges. Biotechnol. Adv. 2019, 37, 109–131. [Google Scholar] [CrossRef]
- Allyn, M.M.; Luo, R.H.; Hellwarth, E.B.; Swindle-Reilly, K.E. Considerations for Polymers Used in Ocular Drug Delivery. Front. Med. 2021, 8, 787644. [Google Scholar] [CrossRef] [PubMed]
- Rizwan, M.; Peh, G.S.L.; Ang, H.P.; Lwin, N.C.; Adnan, K.; Mehta, J.S.; Tan, W.S.; Yim, E.K.F. Sequentially-Crosslinked Bioactive Hydrogels as Nano-Patterned Substrates with Customizable Stiffness and Degradation for Corneal Tissue Engineering Applications. Biomaterials 2017, 120, 139–154. [Google Scholar] [CrossRef]
- Procházková, L.; Rodríguez-Muñoz, Y.; Procházka, J.; Wanner, J. Simple Spectrophotometric Method for Determination of Polyvinylalcohol in Different Types of Wastewater. Int. J. Environ. Anal. Chem. 2014, 94, 399–410. [Google Scholar] [CrossRef]
- Phan, C.M.; Walther, H.; Riederer, D.; Lau, C.; Lorenz, K.O.; Subbaraman, L.N.; Jones, L. Analysis of Polyvinyl Alcohol Release from Commercially Available Daily Disposable Contact Lenses Using an In Vitro Eye Model. J. Biomed. Mater. Res. B Appl. Biomater. 2019, 107, 1662. [Google Scholar] [CrossRef] [PubMed]
- Kesharwani, P.; Bisht, A.; Alexander, A.; Dave, V.; Sharma, S. Biomedical Applications of Hydrogels in Drug Delivery System: An Update. J. Drug Deliv. Sci. Technol. 2021, 66, 102914. [Google Scholar] [CrossRef]
- Buwalda, S.J.; Vermonden, T.; Hennink, W.E. Hydrogels for Therapeutic Delivery: Current Developments and Future Directions. Biomacromolecules 2017, 18, 316–330. [Google Scholar] [CrossRef] [PubMed]
- Parhi, R. Cross-Linked Hydrogel for Pharmaceutical Applications: A Review. Adv. Pharm. Bull. 2017, 7, 515. [Google Scholar] [CrossRef]
- Gastaldi, M.; Cardano, F.; Zanetti, M.; Viscardi, G.; Barolo, C.; Bordiga, S.; Magdassi, S.; Fin, A.; Roppolo, I. Functional Dyes in Polymeric 3D Printing: Applications and Perspectives. ACS Mater. Lett. 2021, 3, 1–17. [Google Scholar] [CrossRef]
- Trucillo, P. Drug Carriers: A Review on the Most Used Mathematical Models for Drug Release. Processes 2022, 10, 1094. [Google Scholar] [CrossRef]
- Hoare, T.R.; Kohane, D.S. Hydrogels in Drug Delivery: Progress and Challenges. Polymer 2008, 49, 1993–2007. [Google Scholar] [CrossRef]
- Neto, A.I.; Demir, K.; Popova, A.A.; Oliveira, M.B.; Mano, J.F.; Levkin, P.A. Fabrication of Hydrogel Particles of Defined Shapes Using Superhydrophobic-Hydrophilic Micropatterns. Adv. Mater. 2016, 28, 7613–7619. [Google Scholar] [CrossRef] [PubMed]
- Rosenfeld, A.; Oelschlaeger, C.; Thelen, R.; Heissler, S.; Levkin, P.A. Miniaturized High-Throughput Synthesis and Screening of Responsive Hydrogels Using Nanoliter Compartments. Mater. Today Bio 2020, 6, 100053. [Google Scholar] [CrossRef] [PubMed]
- Resende, R.; Silva, A.; Marques, C.S.; Rodrigues Arruda, T.; Teixeira, S.C.; Veloso De Oliveira, T. Biodegradation of Polymers: Stages, Measurement, Standards and Prospects. Macromol 2023, 3, 371–399. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ho, B.; Phan, C.-M.; Garg, P.; Shokrollahi, P.; Jones, L. A Rapid Screening Platform for Simultaneous Evaluation of Biodegradation and Therapeutic Release of an Ocular Hydrogel. Pharmaceutics 2023, 15, 2625. https://doi.org/10.3390/pharmaceutics15112625
Ho B, Phan C-M, Garg P, Shokrollahi P, Jones L. A Rapid Screening Platform for Simultaneous Evaluation of Biodegradation and Therapeutic Release of an Ocular Hydrogel. Pharmaceutics. 2023; 15(11):2625. https://doi.org/10.3390/pharmaceutics15112625
Chicago/Turabian StyleHo, Brandon, Chau-Minh Phan, Piyush Garg, Parvin Shokrollahi, and Lyndon Jones. 2023. "A Rapid Screening Platform for Simultaneous Evaluation of Biodegradation and Therapeutic Release of an Ocular Hydrogel" Pharmaceutics 15, no. 11: 2625. https://doi.org/10.3390/pharmaceutics15112625
APA StyleHo, B., Phan, C.-M., Garg, P., Shokrollahi, P., & Jones, L. (2023). A Rapid Screening Platform for Simultaneous Evaluation of Biodegradation and Therapeutic Release of an Ocular Hydrogel. Pharmaceutics, 15(11), 2625. https://doi.org/10.3390/pharmaceutics15112625