Silk Fibroin Bioink for 3D Printing in Tissue Regeneration: Controlled Release of MSC extracellular Vesicles
Abstract
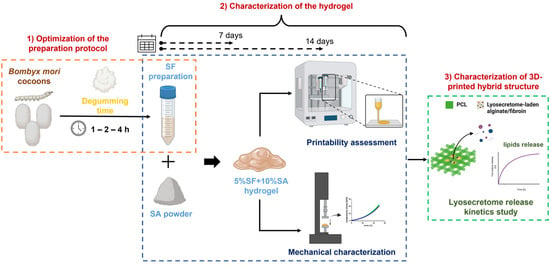
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Silk Fibroin Preparation and Characterization
2.2.1. Silk Fibroin Extraction and Solubilization
2.2.2. Size Exclusion Chromatography (SEC)-UV Analysis
2.2.3. Physico-Chemical Characterization of SF
2.3. Hydrogels and Crosslinking Solution Preparation
2.4. Assessment of SA-SF Hydrogel Printability and Shape Fidelity
2.4.1. 3D Bioprinter and Printing Process
2.4.2. Definition of Printing Parameters and Shape Fidelity Assessment
Serpentine-Like Structure
Grid Structure
2.5. Hydrogel Mechanical Characterization
2.5.1. Sample Preparation
2.5.2. Mechanical Tests
Compression
Tension
2.5.3. Data Elaboration
2.6. Release Study of Lyosecretome
2.6.1. Scaffold Fabrication
2.6.2. Drug Release Studies
2.6.3. Drug Release Kinetic Study
2.7. Statistical Analysis
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wu, C.S.; Wang, B.; Zhang, C.; Wysk, R.A.; Chen, Y.W. Bioprinting: An assessment based on manufacturing readiness levels. Crit. Rev. Biotechnol. 2017, 37, 333–354. [Google Scholar] [CrossRef] [PubMed]
- Mani, M.P.; Sadia, M.; Jaganathan, S.K.; Khudzari, A.Z.; Supriyanto, E.; Saidin, S.; Ramakrishna, S.; Ismail, A.F.; Faudzi, A.A.M. A review on 3D printing in tissue engineering applications. J. Polym. Eng. 2022, 42, 243–265. [Google Scholar] [CrossRef]
- Chia, H.N.; Wu, B.M. Recent advances in 3D printing of biomaterials. J. Biol. Eng. 2015, 9, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melchels, F.P.W.; Feijen, J.; Grijpma, D.W. A review on stereolithography and its applications in biomedical engineering. Biomaterials 2010, 31, 6121–6130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhushan, B.; Caspers, M. An overview of additive manufacturing (3D printing) for microfabrication. Microsyst. Technol. Micro Nanosyst. Inf. Storage Process. Syst. 2017, 23, 1117–1124. [Google Scholar] [CrossRef]
- Rasheed, S.; Lughmani, W.A.; Obeidi, M.A.; Brabazon, D.; Ul Ahad, I. Additive Manufacturing of Bone Scaffolds Using PolyJet and Stereolithography Techniques. Appl. Sci. 2021, 11, 7336. [Google Scholar] [CrossRef]
- Kantaros, A. 3D Printing in Regenerative Medicine: Technologies and Resources Utilized. Int. J. Mol. Sci. 2022, 23, 4621. [Google Scholar] [CrossRef]
- Kantaros, A. Bio-Inspired Materials: Exhibited Characteristics and Integration Degree in Bio-Printing Operations. Am. J. Eng. Appl. Sci. 2022, 15, 255–263. [Google Scholar] [CrossRef]
- De Pieri, A.; Rochev, Y.; Zeugolis, D.I. Scaffold-free cell-based tissue engineering therapies: Advances, shortfalls and forecast. npj Regen. Med. 2021, 6, 18. [Google Scholar] [CrossRef]
- Simunovic, F.; Finkenzeller, G. Vascularization Strategies in Bone Tissue Engineering. Cells 2021, 10, 1749. [Google Scholar] [CrossRef]
- Masson-Meyers, D.S.; Tayebi, L. Vascularization strategies in tissue engineering approaches for soft tissue repair. J. Tissue Eng. Regen. Med. 2021, 15, 747–762. [Google Scholar] [CrossRef]
- Chang, B.; Ahuja, N.; Ma, C.; Liu, X.H. Injectable scaffolds: Preparation and application in dental and craniofacial regeneration. Mater. Sci. Eng. R Rep. 2017, 111, 1–26. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, J.N.; Rungarunlert, S.; Urkasemsin, G.; Adine, C.; Souza, G.R. Three-Dimensional Bioprinting Nanotechnologies towards Clinical Application of Stem Cells and Their Secretome in Salivary Gland Regeneration. Stem Cells Int. 2016, 2016, 7564689. [Google Scholar] [CrossRef] [Green Version]
- Ibrahim, R.; Mndlovu, H.; Kumar, P.; Adeyemi, S.A.; Choonara, Y.E. Cell Secretome Strategies for Controlled Drug Delivery and Wound-Healing Applications. Polymers 2022, 14, 2929. [Google Scholar] [CrossRef]
- Bakopoulou, A.; Huang, G.T.J.; Rouabhia, M.; Geurtsen, W.; About, I. Advances and New Technologies towards Clinical Application of Oral Stem Cells and Their Secretome. Stem Cells Int. 2017, 2017, 6367375. [Google Scholar] [CrossRef]
- Bari, E.; Scocozza, F.; Perteghella, S.; Segale, L.; Sorlini, M.; Auricchio, F.; Conti, M.; Torre, M.L. Three-Dimensional Bioprinted Controlled Release Scaffold Containing Mesenchymal Stem/Stromal Lyosecretome for Bone Regeneration: Sterile Manufacturing and In Vitro Biological Efficacy. Biomedicines 2022, 10, 1063. [Google Scholar] [CrossRef]
- Wechsler, M.E.; Rao, V.V.; Borelli, A.N.; Anseth, K.S. Engineering the MSC Secretome: A Hydrogel Focused Approach. Adv. Healthc. Mater. 2021, 10, 2001948. [Google Scholar] [CrossRef]
- Diomede, F.; D’Aurora, M.; Gugliandolo, A.; Merciaro, I.; Orsini, T.; Gatta, V.; Piattelli, A.; Trubiani, O.; Mazzon, E. Biofunctionalized Scaffold in Bone Tissue Repair. Int. J. Mol. Sci. 2018, 19, 1022. [Google Scholar] [CrossRef] [Green Version]
- Bari, E.; Di Silvestre, D.; Mastracci, L.; Grillo, F.; Grisoli, P.; Marrubini, G.; Nardini, M.; Mastrogiacomo, M.; Sorlini, M.; Rossi, R.; et al. GMP-compliant sponge-like dressing containing MSC lyo-secretome: Proteomic network of healing in a murine wound model. Eur. J. Pharm. Biopharm. 2020, 155, 37–48. [Google Scholar] [CrossRef]
- Lee, K.; Silva, E.A.; Mooney, D.J. Growth factor delivery-based tissue engineering: General approaches and a review of recent developments. J. R. Soc. Interface 2011, 8, 153–170. [Google Scholar] [CrossRef] [Green Version]
- Li, J.Y.; Mooney, D.J. Designing hydrogels for controlled drug delivery. Nat. Rev. Mater. 2016, 1, 16071. [Google Scholar] [CrossRef] [PubMed]
- Su, J.; Hu, B.H.; Lowe, W.L.; Kaufman, D.B.; Messersmith, P.B. Anti-inflammatory peptide-functionalized hydrogels for insulin-secreting cell encapsulation. Biomaterials 2010, 31, 308–314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ganji, F.; Vasheghani-Farahani, E. Hydrogels in Controlled Drug Delivery Systems. Iran. Polym. J. 2009, 18, 63–88. [Google Scholar]
- Rizzo, F.; Kehr, N.S. Recent Advances in Injectable Hydrogels for Controlled and Local Drug Delivery. Adv. Healthc. Mater. 2021, 10, 2001341. [Google Scholar] [CrossRef]
- Bordbar-Khiabani, A.; Gasik, M. Smart Hydrogels for Advanced Drug Delivery Systems. Int. J. Mol. Sci. 2022, 23, 3665. [Google Scholar] [CrossRef]
- Tran, T.S.; Balu, R.; Mettu, S.; Choudhury, N.R.; Dutta, N.K. 4D Printing of Hydrogels: Innovation in Material Design and Emerging Smart Systems for Drug Delivery. Pharmaceuticals 2022, 15, 1282. [Google Scholar] [CrossRef]
- Bari, E.; Scocozza, F.; Perteghella, S.; Sorlini, M.; Auricchio, F.; Torre, M.L.; Conti, M. 3D Bioprinted Scaffolds Containing Mesenchymal Stem/Stromal Lyosecretome: Next Generation Controlled Release Device for Bone Regenerative Medicine. Pharmaceutics 2021, 13, 515. [Google Scholar] [CrossRef]
- Jiao, W.J.; Li, X.H.; Shan, J.X.; Wang, X.H. Study of Several Alginate-Based Hydrogels for In Vitro 3D Cell Cultures. Gels 2022, 8, 147. [Google Scholar] [CrossRef]
- Lu, Q.; Zhu, H.; Zhang, C.; Zhang, F.; Zhang, B.; Kaplan, D.L. Silk Self-Assembly Mechanisms and Control From Thermodynamics to Kinetics. Biomacromolecules 2012, 13, 826–832. [Google Scholar] [CrossRef] [Green Version]
- Meinel, L.; Hofmann, S.; Karageorgiou, V.; Kirker-Head, C.; McCool, J.; Gronowicz, G.; Zichner, L.; Langer, R.; Vunjak-Novakovic, G.; Kaplan, D.L. The inflammatory responses to silk films in vitro and in vivo. Biomaterials 2005, 26, 147–155. [Google Scholar] [CrossRef]
- Santin, M.; Motta, A.; Freddi, G.; Cannas, M. In vitro evaluation of the inflammatory potential of the silk fibroin. J. Biomed. Mater. Res. 1999, 46, 382–389. [Google Scholar] [CrossRef]
- Panilaitis, B.; Altman, G.H.; Chen, J.S.; Jin, H.J.; Karageorgiou, V.; Kaplan, D.L. Macrophage responses to silk. Biomaterials 2003, 24, 3079–3085. [Google Scholar] [CrossRef]
- Vepari, C.; Kaplan, D.L. Silk as a biomaterial. Prog. Polym. Sci. 2007, 32, 991–1007. [Google Scholar] [CrossRef]
- Liu, T.-l.; Miao, J.-c.; Sheng, W.-h.; Xie, Y.-f.; Huang, Q.; Shan, Y.-b.; Yang, J.-c. Cytocompatibility of regenerated silk fibroin film: A medical biomaterial applicable to wound healing. J. Zhejiang Univ. Sci. B 2010, 11, 10–16. [Google Scholar] [CrossRef] [Green Version]
- Horan, R.L.; Antle, K.; Collette, A.L.; Huang, Y.Z.; Huang, J.; Moreau, J.E.; Volloch, V.; Kaplan, D.L.; Altman, G.H. In vitro degradation of silk fibroin. Biomaterials 2005, 26, 3385–3393. [Google Scholar] [CrossRef]
- Kim, D.W.; Hwang, H.S.; Kim, D.S.; Sheen, S.H.; Heo, D.H.; Hwang, G.; Kang, S.H.; Kweon, H.; Jo, Y.Y.; Kang, S.W.; et al. Effect of silk fibroin peptide derived from silkworm Bombyx mori on the anti-inflammatory effect of Tat-SOD in a mice edema model. BMB Rep. 2011, 44, 787–792. [Google Scholar] [CrossRef] [Green Version]
- Lozano-Perez, A.A.; Rodriguez-Nogales, A.; Ortiz-Cullera, V.; Algieri, F.; Garrido-Mesa, J.; Zorrilla, P.; Rodriguez-Cabezas, M.E.; Garrido-Mesa, N.; Utrilla, M.P.; De Matteis, L.; et al. Silk fibroin nanoparticles constitute a vector for controlled release of resveratrol in an experimental model of inflammatory bowel disease in rats. Int. J. Nanomed. 2014, 9, 4507–4520. [Google Scholar] [CrossRef] [Green Version]
- Crivelli, B.; Bari, E.; Perteghella, S.; Catenacci, L.; Sorrenti, M.; Mocchi, M.; Farago, S.; Tripodo, G.; Prina-Mello, A.; Torre, M.L. Silk fibroin nanoparticles for celecoxib and curcumin delivery: ROS-scavenging and anti-inflammatory activities in an in vitro model of osteoarthritis. Eur. J. Pharm. Biopharm. 2019, 137, 37–45. [Google Scholar] [CrossRef]
- Ayoub, N.A.; Garb, J.E.; Tinghitella, R.M.; Collin, M.A.; Hayashi, C.Y. Blueprint for a High-Performance Biomaterial: Full-Length Spider Dragline Silk Genes. PLoS One 2007, 2, e514. [Google Scholar] [CrossRef]
- Jin, H.J.; Kaplan, D.L. Mechanism of silk processing in insects and spiders. Nature 2003, 424, 1057–1061. [Google Scholar] [CrossRef]
- Matsumoto, A.; Lindsay, A.; Abedian, B.; Kaplan, D.L. Silk Fibroin Solution Properties Related to Assembly and Structure. Macromol. Biosci. 2008, 8, 1006–1018. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Shao, Z.; Knight, D.P.; Vollrath, F. Conformation transition kinetics of Bombyx mori silk protein. Proteins-Struct. Funct. Bioinform. 2007, 68, 223–231. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.S.; Han, G.C.; Yan, S.Q.; Zhang, Q. 3D Printing of Silk Fibroin for Biomedical Applications. Materials 2019, 12, 504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wlodarczyk-Biegun, M.K.; del Campo, A. 3D bioprinting of structural proteins. Biomaterials 2017, 134, 180–201. [Google Scholar] [CrossRef]
- DeSimone, E.; Schacht, K.; Jungst, T.; Groll, J.; Scheibel, T. Biofabrication of 3D constructs: Fabrication technologies and spider silk proteins as bioinks. Pure Appl. Chem. 2015, 87, 737–749. [Google Scholar] [CrossRef] [Green Version]
- Rockwood, D.N.; Preda, R.C.; Yucel, T.; Wang, X.; Lovett, M.L.; Kaplan, D.L. Materials fabrication from Bombyx mori silk fibroin. Nat. Protoc. 2011, 6, 1612–1631. [Google Scholar] [CrossRef] [Green Version]
- Phillips, D.M.; Drummy, L.F.; Conrady, D.G.; Fox, D.M.; Naik, R.R.; Stone, M.O.; Trulove, P.C.; De Long, H.C.; Mantz, R.A. Dissolution and regeneration of Bombyx mori Silk fibroin using ionic liquids. J. Am. Chem. Soc. 2004, 126, 14350–14351. [Google Scholar] [CrossRef]
- Tengattini, S.; Orlandi, G.; Perteghella, S.; Bari, E.; Amadio, M.; Calleri, E.; Massolini, G.; Torre, M.L.; Temporini, C. Chromatographic profiling of silk sericin for biomedical and cosmetic use by complementary hydrophylic, reversed phase and size exclusion chromatographic methods. J. Pharm. Biomed. Anal. 2020, 186, 113291. [Google Scholar] [CrossRef]
- Scocozza, F.; Sakaj, M.; Auricchio, F.; Marconi, S.; Riello, P.; Ferrari, C.; Cansolino, L.; Catenacci, L.; Sorrenti, M.; Scatto, M.; et al. Shape fidelity and sterility assessment of 3D printed polycaprolactone and hydroxyapatite scaffolds. J. Polym. Res. 2021, 28, 327. [Google Scholar] [CrossRef]
- Kantaros, A.; Dimitrios, P. Employing a Low-Cost Desktop 3D Printer: Challenges, and How to Overcome Them by Tuning Key Process Parameters. Int. J. Mech. Appl. 2021, 10, 11–19. [Google Scholar] [CrossRef]
- Paxton, N.; Smolan, W.; Bock, T.; Melchels, F.; Groll, J.; Jungst, T. Proposal to assess printability of bioinks for extrusion-based bioprinting and evaluation of rheological properties governing bioprintability. Biofabrication 2017, 9, 044107. [Google Scholar] [CrossRef] [Green Version]
- Schwab, A.; Levato, R.; D’Este, M.; Piluso, S.; Eglin, D.; Malda, J. Printability and Shape Fidelity of Bioinks in 3D Bioprinting. Chem. Rev. 2020, 120, 10850–10877. [Google Scholar] [CrossRef]
- Ragazzini, S.; Scocozza, F.; Bernava, G.; Auricchio, F.; Colombo, G.I.; Barbuto, M.; Conti, M.; Pesce, M.; Garoffolo, G. Mechanosensor YAP cooperates with TGF- beta 1 signaling to promote myofibroblast activation and matrix stiffening in a 3D model of human cardiac fibrosis. Acta Biomater. 2022, 152, 300–312. [Google Scholar] [CrossRef]
- Baniasadi, M.; Minary-Jolandan, M. Alginate-Collagen Fibril Composite Hydrogel. Materials 2015, 8, 799–814. [Google Scholar] [CrossRef] [Green Version]
- Xu, C.C.; Lee, W.H.; Dai, G.H.; Hong, Y. Highly Elastic Biodegradable Single-Network Hydrogel for Cell Printing. ACS Appl. Mater. Interfaces 2018, 10, 9969–9979. [Google Scholar] [CrossRef]
- Di Giuseppe, M.; Law, N.; Webb, B.; Macrae, R.A.; Liew, L.J.; Sercombe, T.B.; Dilley, R.J.; Doyle, B.J. Mechanical behaviour of alginate-gelatin hydrogels for 3D bioprinting. J. Mech. Behav. Biomed. Mater. 2018, 79, 150–157. [Google Scholar] [CrossRef]
- Bari, E.; Perteghella, S.; Di Silvestre, D.; Sorlini, M.; Catenacci, L.; Sorrenti, M.; Marrubini, G.; Rossi, R.; Tripodo, G.; Mauri, P.; et al. Pilot Production of Mesenchymal Stem/Stromal Freeze-Dried Secretome for Cell-Free Regenerative Nanomedicine: A Validated GMP-Compliant Process. Cells 2018, 7, 190. [Google Scholar] [CrossRef] [Green Version]
- Caccavo, D. An overview on the mathematical modeling of hydrogels’ behavior for drug delivery systems. Int. J. Pharm. 2019, 560, 175–190. [Google Scholar] [CrossRef]
- Gulrajani, M.L.; Agarwal, R.; Grover, A.; Suri, M. Degumming of silk with lipase and protease. Indian J. Fibre Text. Res. 2000, 25, 69–74. [Google Scholar]
- Chang, H.P.; Cheng, T.C.; Wu, Y.Q.; Hu, W.B.; Long, R.W.; Liu, C.; Zhao, P.; Xia, Q.Y. Transcriptomic Analysis of the Anterior Silk Gland in the Domestic Silkworm (Bombyx mori)—Insight into the Mechanism of Silk Formation and Spinning. PLoS One 2015, 10, e0139424. [Google Scholar] [CrossRef] [Green Version]
- Ling, S.J.; Qi, Z.M.; Knight, D.P.; Shao, Z.Z.; Chen, X. Synchrotron FTIR Microspectroscopy of Single Natural Silk Fibers. Biomacromolecules 2011, 12, 3344–3349. [Google Scholar] [CrossRef] [PubMed]
- Um, I.C.; Kweon, H.Y.; Lee, K.G.; Park, Y.H. The role of formic acid in solution stability and crystallization of silk protein polymer. Int. J. Biol. Macromol. 2003, 33, 203–213. [Google Scholar] [CrossRef] [PubMed]
- Kuo, C.K.; Ma, P.X. Ionically crosslinked alginate hydrogels as scaffolds for tissue engineering: Part 1. Structure, gelation rate and mechanical properties. Biomaterials 2001, 22, 511–521. [Google Scholar] [CrossRef] [PubMed]
- Silva, R.; Singh, R.; Sarker, B.; Papageorgiou, D.G.; Juhasz, J.A.; Roether, J.A.; Cicha, I.; Kaschta, J.; Schubert, D.W.; Chrissafis, K.; et al. Soft-matrices based on silk fibroin and alginate for tissue engineering. Int. J. Biol. Macromol. 2016, 93, 1420–1431. [Google Scholar] [CrossRef]
- Adi, A.; Haim, S.M.; Smadar, E.S.; Mirit, S. The mechanical behavior of silk-fibroin reinforced alginate hydrogel biocomposites—Toward functional tissue biomimetics. J. Mech. Behav. Biomed. Mater. 2023, 138, 105598. [Google Scholar]
- Peppas, N.A.; Narasimhan, B. Mathematical models in drug delivery: How modeling has shaped the way we design new drug delivery systems. J. Control. Release 2014, 190, 75–81. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhu, Z.S.; Guan, J.; Wu, S.J. Processing, mechanical properties and bio-applications of silk fibroin-based high-strength hydrogels. Acta Biomater. 2021, 125, 57–71. [Google Scholar] [CrossRef]
- Peppas, N.A. Analysis of Fickian and Non-Fickian Drug Release from Polymers. Pharm. Acta Helv. 1985, 60, 110–111. [Google Scholar]






| Process | Solvent | Temperature | Time |
|---|---|---|---|
| Standard | Na2CO3 0.02 M | 100 °C | 30 min |
| Short alkaline boiling | 1 h | ||
| Medium alkaline boiling | 2 h | ||
| Intensive alkaline boiling | 4 h |
| Formulation | Formulation (w/v) | SF Degumming Time (h) |
|---|---|---|
| SA-SF-1 h | SA 10% + SF 5% | 1 |
| SA-SF-2 h | 2 | |
| SA-SF-4 h | 4 | |
| SA (CRTL) | SA 10% | / |
| Hydrogel | Condition | Compression | Tensile |
|---|---|---|---|
| SA-SF-1h | t1 (7 days) | 4 | 4 |
| t2 (14 days) | 6 | 5 | |
| SA-SF-2h | t1 (7 days) | 6 | 4 |
| t2 (14 days) | 6 | 5 | |
| SA-SF-4h | t1 (7 days) | 6 | 2 |
| t2 (14 days) | 6 | 4 | |
| SA (CTRL) | CaCl2 + KCl + protamine | 6 | 5 |
| CaCl2 | - | 6 |
| Degumming Time (h) | Elution Volume ± SD (mL) | Estimated MW (Da) | Area ± SD | Area RSD (%) |
|---|---|---|---|---|
| 0.5 | 2.724 ± 0.04 | > 500 * | 2011 ± 105 | 5.2 |
| 1 | 3.186 ± 0.05 | 174 | 2317 ± 75 | 3.3 |
| 2 | 3.581 ± 0.05 | 60 | 2235 ± 59 | 2.6 |
| 3 | 3.392 ± 0.05 | 24 | 2271 ± 18 | 0.8 |
| Hydrogel | Printing Parameters | t1 (7 Days) | t2 (14 Days) | ||
|---|---|---|---|---|---|
| Nozzle 0.41 mm | Nozzle 0.25 mm | Nozzle 0.41 mm | Nozzle 0.25 mm | ||
| SA-SF-1h | Speed (mm/min) | 1000 | 1000 | n.p. | n.p. |
| Pressure (kPa) | 8 | 12 | n.p. | n.p. | |
| SA-SF-2h | Speed (mm/min) | 600 | 600 | 600 | 600 |
| Pressure (kPa) | 20 | 30 | 20 | 30 | |
| SA-SF-4h | Speed (mm/min) | 600 | 600 | 600 | 600 |
| Pressure (kPa) | 20 | 35 | 20 | 35 | |
| Hydrogel | Nozzle (mm) | t1 (7 Days) | t2 (14 Days) | ||||
|---|---|---|---|---|---|---|---|
| PA (%) Filament Width | PA (%) Inter-Filament Distance | Pr | PA (%) Filament Width | PA (%) Inter-Filament Distance | Pr | ||
| SA-SF-1h | 0.41 | 51.6 | 79 | 0.83 | / | / | / |
| 0.25 | 17.7 | 63 | 0.9 | / | / | / | |
| SA-SF-2h | 0.41 | 90 | 93 | 0.88 | 39.5 | 56.5 | 0.9 |
| 0.25 | 25.7 | 90.3 | 0.91 | 9 | 65.4 | 0.87 | |
| SA-SF-4h | 0.41 | 93.4 | 58 | 0.85 | 76.2 | 90 | 0.85 |
| 0.25 | 60 | 91.4 | 0.86 | 16.3 | 66 | 0.85 | |
| Model | Equation | Formulation | SF Degumming Time (h) | Coefficients (95% Confidence Bounds) | Sum of Squares | R2 | Degrees of Freedom |
|---|---|---|---|---|---|---|---|
| Higuchi | F(t) = k × t0.5 | SF + alginate | 1 | k = 3.192 (2.525, 3.859) | 7317 | 0.217 | 21 |
| 2 | k = 1.919 (1.776, 2.061) | 334.3 | 0.8972 | 21 | |||
| 4 | k = 2.179 (1.996, 2.362) | 550.9 | 0.9012 | 21 | |||
| Alginate | / | k = 1.394 (1.188, 1.600) | 698.5 | 0.3735 | 21 | ||
| Higuchi (eq 2.12 from [58]) | F(t) = 100 × (1 − C × exp (−k × t)) | SF + alginate | 1 | C = 0.6746 (0.6237, 0.7260) k = 0.00107 (0.0006436, 0.001540) | 0.1290 | 0.6093 | 22 |
| 2 | C = 0.9037 (0.8825, 0.9250) k = 0.0009412 (0.0008108, 0.001075) | 227.1 | 0.9301 | 22 | |||
| 4 | C = 0.9232 (0.8952, 0.9513) k = 0.001269 (0.001088, 0.001457) | 389.7 | 0.9301 | 22 | |||
| Alginate | / | C = 0.887 (0.8605, 0.9137) k = 0.000429 (0.0002833, 0.0005791) | 372.4 | 0.6659 | 22 | ||
| Peppas– Sahlin | F(t) = k1 × tm + k2 × t(2 × m) | SF + alginate | 1 | k1 = ~ 21.45 k2 = ~ 8.173 M = ~ 0.07298 | 115.1 | 0.9651 | 22 |
| 2 | k1 = ~ 4.311 k2 = ~ 0.6770 M = ~ 0.2624 | 137.7 | 0.9576 | 22 | |||
| 4 | k1 = ~ 2.304 k2 = ~ 0.1857 M = ~ 0.3911 | 461.5 | 0.9173 | 22 | |||
| Alginate | / | k1 = -0.1679 (-0.3920, -0.01953) k2 = 0.2406 (0.1669, 0.3539) m = 0.2023 (0.06861, 0.4205) | 0.01727 | 0.9706 | 22 | ||
| Ritger– Peppas | F(t) = k × tn | SF + alginate | 1 | k = 29.56 (27.43, 31.73) n = 0.09653 (0.08189, 0.1117) | 121.1 | 0.9633 | 22 |
| 2 | k = 4.572 (3.189, 6.214) n = 0.3454 (0.2892, 0.4099) | 163.9 | 0.9496 | 22 | |||
| 4 | k = 1.982 (0.7195, 4.194) n = 0.5166 (0.3834, 0.6913) | 549.4 | 0.9015 | 22 | |||
| Alginate | / | k = 7.767 (6.967, 8.600) n = 0.1922 (0.1725, 0.2128) | 26.66 | 0.9761 | 22 | ||
| Zero-order | F(t) = k × t | SF + alginate | 1 | k = 0.1509 (0.09954, 0.2023) | 15065 | 0.3564 | 22 |
| 2 | k = 0.09762 (0.08080, 0.1144) | 1613 | 0.5038 | 22 | |||
| 4 | k = 0.1145 (0.1001, 0.1291) | 1200 | 0.7849 | 22 | |||
| Alginate | / | k = 0.06667 (0.04768, 0.08566) | 2057 | 0.2161 | 22 | ||
| Korsmeyer– Peppas | F(t) = kKP × tn × Q0 | SF + alginate | 1 | kKP = 29.56 (27.43, 31.73) n = 0.09653 (0.08189, 0.1117) | 121.1 | 0.9633 | 22 |
| 2 | kKP = 4.572 (3.189, 6.214) n = 0.3454 (0.2892, 0.4099) | 163.9 | 0.9496 | 22 | |||
| 4 | kKP = 1.982 (0.7195, 4.194) n = 0.5166 (0.3834, 0.6913) | 549.4 | 0.9015 | 22 | |||
| Alginate | / | kKP = 7.767 (6.967, 8.600) n = 0.1922 (0.1725, 0.2128) | 26.66 | 0.9761 | 22 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bari, E.; Di Gravina, G.M.; Scocozza, F.; Perteghella, S.; Frongia, B.; Tengattini, S.; Segale, L.; Torre, M.L.; Conti, M. Silk Fibroin Bioink for 3D Printing in Tissue Regeneration: Controlled Release of MSC extracellular Vesicles. Pharmaceutics 2023, 15, 383. https://doi.org/10.3390/pharmaceutics15020383
Bari E, Di Gravina GM, Scocozza F, Perteghella S, Frongia B, Tengattini S, Segale L, Torre ML, Conti M. Silk Fibroin Bioink for 3D Printing in Tissue Regeneration: Controlled Release of MSC extracellular Vesicles. Pharmaceutics. 2023; 15(2):383. https://doi.org/10.3390/pharmaceutics15020383
Chicago/Turabian StyleBari, Elia, Giulia Maria Di Gravina, Franca Scocozza, Sara Perteghella, Benedetta Frongia, Sara Tengattini, Lorena Segale, Maria Luisa Torre, and Michele Conti. 2023. "Silk Fibroin Bioink for 3D Printing in Tissue Regeneration: Controlled Release of MSC extracellular Vesicles" Pharmaceutics 15, no. 2: 383. https://doi.org/10.3390/pharmaceutics15020383
APA StyleBari, E., Di Gravina, G. M., Scocozza, F., Perteghella, S., Frongia, B., Tengattini, S., Segale, L., Torre, M. L., & Conti, M. (2023). Silk Fibroin Bioink for 3D Printing in Tissue Regeneration: Controlled Release of MSC extracellular Vesicles. Pharmaceutics, 15(2), 383. https://doi.org/10.3390/pharmaceutics15020383