3.1. mRNA Transfection into Immune Cells
Six previously reported modified mRNA formulations were evaluated in this study [
3,
8,
19,
20]: (1) 10% 5mC, 10% 2sU; (2) 25% 5mC, 25% 2sU; (3) 20% 5mC, 10% 2sU, 10% ψU; (4) 100% 5mC; (5) 100% ψU; (6) 100% 5mC, 100% ψU. The percentages for each formulation indicate the percentage of total cytidine or uridine in mRNA that is a modified cytidine or uridine analog. These formulations will be abbreviated as (1) mC
10% sU
10%, (2) mC
25% sU
25%, (3) mC
20% sU
10% ψU
10%, (4) mC
100%, (5) ψU
100% and (6) mC
100% ψU
100%.
The immunogenicity of each formulation was evaluated by mRNA transfection into RAW264.7 cells (a mouse leukemic monocyte macrophage cell line). The modified mRNA encoded the reporter
Gaussia luciferase (GLuc), a secretable form of luciferase [
21]. In this experiment, we used serum-free medium with the intention to focus on the effect of mRNA immunogenicity by excluding the effect of the other factors such as degradation in the cultured medium. Four hours after mRNA transfection using Lipofectamine™ LTX, the expression of the inflammatory factors interferon-β (IFN-β), interleukin-6 (IL-6), tumor necrosis factor-α (TNF-α), and interleukin-1β (IL-1β) was measured by qRT-PCR.
The expression of IFN-β and IL-6 increased remarkably after transfection of unmodified mRNA compared with the untransfected control (
Figure 1). For five of the modified mRNA formulations, except for ψU
100%, the expression showed only a mild increase from the untransfected control. Among the modification formulations, the mC
100% and the mC
100% ψU
100% formulations had the most reduced inflammatory responses. In addition, cell viability was evaluated at the same time point. While transfection of unmodified mRNA caused some decrease in cell viability, the use of modified mRNAs generally alleviated the cytotoxicity (
Figure S1). By making scatter plots representing the correlation between cytokine induction and cell viability after mRNA transfection (
Figure S2), it is well documented that the cell viability inversely correlated with the cytokine production with the exception of ψU
100%.
The protein expression efficiency for each formulation was determined by measuring GLuc secreted into the culture media. The protein expression efficiency was then compared with the IFN-β and IL-6 response induced by each formulation. Four hours after transfection, GLuc expression was significantly increased for the mC
100% and the mC
100% ψU
100% formulations while the other four formulations had GLuc expression levels similar to that of the unmodified mRNA (
Figure 2a). mRNA with mC
100% and mC
100% ψU
100% exhibited high transgene expression with less induction of an inflammatory response (IFN-β or IL-6) (
Figure 2b,c). Of the formulations tested, these two are likely optimal for obtaining high protein yield with low immunogenicity. In contrast, the mC
10% sU
10%, mC
25% sU
25%, and mC
20% sU
10% ψU
10% formulations have reduced immunogenicity, but they did not show significant increase in luciferase expression compared to that of the unmodified mRNA. mRNA with ψU
100% resulted in GLuc expression comparable to unmodified mRNA, but it induced a much stronger inflammatory response. In accordance with these results, although cell viability was positively correlated with luciferase expression efficiency, some formulations, especially mC
25% sU
25%, exhibited only a small increase in luciferase expression despite remarkable increase in cell viability (
Figure S2). These results indicate that the reduced inflammatory responses, as well as the increase in cell viability, are not always correlated with an increase in protein expression efficiency, motivating us to analyze the effect of mRNA modification using other cell types.
Figure 1.
Inflammatory response after mRNA introduction into RAW264.7 cells. mRNA-encoding GLuc was introduced to RAW264.7 cells using Lipofectamine™ LTX. After 4 h, expression of inflammatory molecules was measured with qRT-PCR. (a) Interferon-β (IFN-β); (b) Interleukin-6 (IL-6); (c) Tumor necrosis factor-α (TNF-α); and (d) Interleukin-1β (IL-1β). Data are presented as the mean ± standard error of the mean (S.E.M.) (N = 6) for expression levels relative to the untransfected control.
Figure 1.
Inflammatory response after mRNA introduction into RAW264.7 cells. mRNA-encoding GLuc was introduced to RAW264.7 cells using Lipofectamine™ LTX. After 4 h, expression of inflammatory molecules was measured with qRT-PCR. (a) Interferon-β (IFN-β); (b) Interleukin-6 (IL-6); (c) Tumor necrosis factor-α (TNF-α); and (d) Interleukin-1β (IL-1β). Data are presented as the mean ± standard error of the mean (S.E.M.) (N = 6) for expression levels relative to the untransfected control.
Figure 2.
GLuc expression in RAW264.7 cells. mRNA-encoding GLuc was introduced to RAW264.7 cells using Lipofectamine™ LTX. After 4 h, the amount of GLuc protein in culture media was measured. (
a) Efficiency of GLuc expression. The graph shows relative values compared to GLuc expression after transfection of unmodified mRNA. Data are presented as the mean ± standard error of the mean (S.E.M.) (
N = 6). Statistical significance was assessed by one-way analysis of variance (ANOVA) followed by Dunnett’s test.
*** p < 0.001
versus unmodified mRNA group. (
b,c) Correlation between inflammatory responses and GLuc expression. The
x-axis shows the expression of (
b) interferon-β (IFN-β) and (
c) interleukin-6 (IL-6) (see
Figure 1). The
y-axis showed the efficiency of GLuc expression (panel (
a) of this figure).
Figure 2.
GLuc expression in RAW264.7 cells. mRNA-encoding GLuc was introduced to RAW264.7 cells using Lipofectamine™ LTX. After 4 h, the amount of GLuc protein in culture media was measured. (
a) Efficiency of GLuc expression. The graph shows relative values compared to GLuc expression after transfection of unmodified mRNA. Data are presented as the mean ± standard error of the mean (S.E.M.) (
N = 6). Statistical significance was assessed by one-way analysis of variance (ANOVA) followed by Dunnett’s test.
*** p < 0.001
versus unmodified mRNA group. (
b,c) Correlation between inflammatory responses and GLuc expression. The
x-axis shows the expression of (
b) interferon-β (IFN-β) and (
c) interleukin-6 (IL-6) (see
Figure 1). The
y-axis showed the efficiency of GLuc expression (panel (
a) of this figure).
3.2. Influence of Transfection Conditions on Protein Expression from mRNA
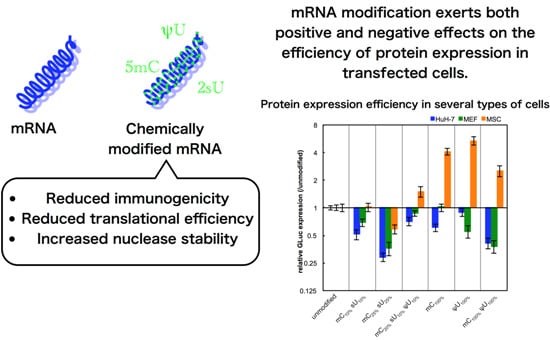
The non-immune cells (cancer-derived cell line (HuH-7), primary cells (MEFs), and adult tissue stem cells (MSCs)) were used for evaluating the effect of mRNA modification on protein expression efficiency. In these experiments, we used serum-containing medium with the intention of evaluating the combinational effect of various factors that can affect
in vitro transfection. Unexpectedly, most of the formulations had reduced GLuc expression in HuH-7 cells and MEFs after 24 h of transfection (
Figure 3). In contrast, in MSCs, modified mRNA generally exhibited higher GLuc expression than the unmodified mRNA. The mC
100% and ψU
100% formulations had the highest GLuc expression in MSCs. Protein production efficiency from modified mRNA depends on cell type.
Figure 3.
GLuc expression in HuH-7 cells, MEFs, and MSCs. mRNA-encoding GLuc was introduced to HuH-7 cells (black bars), MEFs (gray bars), and MSCs (white bars) using Lipofectamine™ LTX. After 24 h, the amount of GLuc protein in culture media was measured. The graph shows relative values compared to GLuc expression after transfection of unmodified mRNA. Data are presented as the mean ± standard error of the mean (S.E.M.) (N = 6). Statistical significance was assessed by one-way analysis of variance (ANOVA) followed by Dunnett’s test. * p < 0.05, ** p < 0.01, *** p < 0.001 versus unmodified mRNA.
Figure 3.
GLuc expression in HuH-7 cells, MEFs, and MSCs. mRNA-encoding GLuc was introduced to HuH-7 cells (black bars), MEFs (gray bars), and MSCs (white bars) using Lipofectamine™ LTX. After 24 h, the amount of GLuc protein in culture media was measured. The graph shows relative values compared to GLuc expression after transfection of unmodified mRNA. Data are presented as the mean ± standard error of the mean (S.E.M.) (N = 6). Statistical significance was assessed by one-way analysis of variance (ANOVA) followed by Dunnett’s test. * p < 0.05, ** p < 0.01, *** p < 0.001 versus unmodified mRNA.
We then evaluated other factors that may affect protein expression efficiency, such as transfection reagents and the gene encoded by the mRNA. When linear polyethyleneimine (LPEI) was used for transfection, the efficiency with the control unmodified mRNA was comparable to that using Lipofectamine™ LTX (
Figure S3). However, the effect of mRNA modification on the protein expression greatly varied with the transfection reagents, particularly in HuH-7 cells (
Figure 4). It is speculated that these complex results may be due to the different intracellular mechanisms between the two reagents. Regarding genes encoded by the mRNA, the expression of firefly luciferase (Luc2) showed trends similar to GLuc (
Figure 5). However, large differences between the two genes were observed in some cases.
Figure 4.
GLuc expression efficiency after mRNA introduction using two different transfection reagents. Two types of transfection reagents, Lipofectamine™ LTX (Lipo, black bars) and linear polyethyleneimine (LPEI, white bars), were used for mRNA introduction. mRNA-encoding GLuc was introduced to (
a) HuH-7, (
b) MEFs, and (
c) MSCs. After 24 h, the amount of GLuc protein in culture media was measured. The graph shows relative values compared to GLuc expression after transfection of unmodified mRNA. For Lipo-treated groups, the same data shown in
Figure 3 are presented. Data are presented as the mean ± standard error of the mean (S.E.M.) (
N = 6). Statistical significance was assessed by one-way analysis of variance (ANOVA) followed by Dunnett’s test.
* p < 0.05,
** p < 0.01,
*** p < 0.001 versus unmodified mRNA.
Figure 4.
GLuc expression efficiency after mRNA introduction using two different transfection reagents. Two types of transfection reagents, Lipofectamine™ LTX (Lipo, black bars) and linear polyethyleneimine (LPEI, white bars), were used for mRNA introduction. mRNA-encoding GLuc was introduced to (
a) HuH-7, (
b) MEFs, and (
c) MSCs. After 24 h, the amount of GLuc protein in culture media was measured. The graph shows relative values compared to GLuc expression after transfection of unmodified mRNA. For Lipo-treated groups, the same data shown in
Figure 3 are presented. Data are presented as the mean ± standard error of the mean (S.E.M.) (
N = 6). Statistical significance was assessed by one-way analysis of variance (ANOVA) followed by Dunnett’s test.
* p < 0.05,
** p < 0.01,
*** p < 0.001 versus unmodified mRNA.
Figure 5.
Influence of mRNA modification on the expression of GLuc and Luc2. Two genes,
Gaussia luciferase (GLuc, black bars) and a synthetic firefly luciferase (Luc2, white bars), were introduced to (
a) HuH-7 cells, (
b) MEFs, and (
c) MSCs using Lipofectamine™ LTX. After 24 h, GLuc protein in culture media and Luc2 protein in cell lysates were measured. The graph shows relative values compared to the expression after transfection of unmodified mRNA. For GLuc-treated cells, the data is the same as in
Figure 3. Data are presented as the mean ± standard error of the mean (S.E.M.) (
N = 6). Statistical significance was assessed by one-way analysis of variance (ANOVA) followed by Dunnett’s test.
* p < 0.05,
** p < 0.01,
*** p < 0.001
versus unmodified mRNA.
Figure 5.
Influence of mRNA modification on the expression of GLuc and Luc2. Two genes,
Gaussia luciferase (GLuc, black bars) and a synthetic firefly luciferase (Luc2, white bars), were introduced to (
a) HuH-7 cells, (
b) MEFs, and (
c) MSCs using Lipofectamine™ LTX. After 24 h, GLuc protein in culture media and Luc2 protein in cell lysates were measured. The graph shows relative values compared to the expression after transfection of unmodified mRNA. For GLuc-treated cells, the data is the same as in
Figure 3. Data are presented as the mean ± standard error of the mean (S.E.M.) (
N = 6). Statistical significance was assessed by one-way analysis of variance (ANOVA) followed by Dunnett’s test.
* p < 0.05,
** p < 0.01,
*** p < 0.001
versus unmodified mRNA.
3.3. Evaluation of Translation Efficiency and Nuclease Stability of Modified mRNA
Although the mRNA modification showed reduced immunogenicity in immune cells, it may cause negative effects on protein expression efficiency in non-immune cells (
Figure 3,
Figure 4 and
Figure 5). To directly analyze protein translation, we used a cell-free translational system based on rabbit reticulocyte lysates containing all molecules that would be required for protein translation from mRNA. Using this system, translation efficiency can be measured independent of other factors such as cellular uptake, endosomal escape, and transport of mRNA to the place of translation.
Translational activity tended to decrease after mRNA modification compared to that of unmodified mRNA, although the extent of the decrease was considerably different depending on the formulation (
Figure 6). The mRNA with mC
100% and mC
100% ψU
100%, which had increased protein expression in Raw 264.7 cells and MSCs (
Figure 2,
Figure 3,
Figure 4 and
Figure 5), showed a large decrease in protein translation in the cell-free system.
Figure 6.
Translation efficiency of modified mRNA. mRNA-encoding GLuc in a naked form was incubated in a cell-free translation system composed of rabbit reticulocyte lysate. After 90 min, GLuc was measured. The graph shows relative values compared to the expression of unmodified mRNA. Data are presented as the mean ± standard error of the mean (S.E.M.) (N = 4). Statistical significance was assessed by one-way analysis of variance (ANOVA) followed by Dunnett’s test. * p < 0.05, *** p < 0.001 versus unmodified mRNA.
Figure 6.
Translation efficiency of modified mRNA. mRNA-encoding GLuc in a naked form was incubated in a cell-free translation system composed of rabbit reticulocyte lysate. After 90 min, GLuc was measured. The graph shows relative values compared to the expression of unmodified mRNA. Data are presented as the mean ± standard error of the mean (S.E.M.) (N = 4). Statistical significance was assessed by one-way analysis of variance (ANOVA) followed by Dunnett’s test. * p < 0.05, *** p < 0.001 versus unmodified mRNA.
Another factor that might affect protein expression is the nuclease stability of mRNA under physiological conditions. To evaluate this, the modified mRNA formulations were incubated in naked form in 0.05% serum for 15 min at 37 °C. The amount of preserved mRNA was quantified by qRT-PCR. The modified mRNA generally showed enhanced stability compared with unmodified mRNA (
Figure 7). In particular, the mRNA containing ψU
100% or mC
100% ψU
100% exhibited significantly higher stability, corresponding to the high protein expression detected in Raw 264.7 cells or MSCs (
Figure 2,
Figure 3,
Figure 4 and
Figure 5).
Figure 7.
Nuclease stability of mRNA. mRNA-encoding GLuc in a naked form was incubated in 0.05% serum at 37 °C for 15 min. qRT-PCR measurement was performed to quantify the amount of mRNA that has an intact sequence between a chosen primer pair. The amount of mRNA after serum incubation was compared to that before serum incubation and the relative values are shown in the graph. Data are presented as the mean ± standard error of the mean (S.E.M.) (N = 4). Statistical significance was assessed by one-way analysis of variance (ANOVA) followed by Dunnett’s test. * p < 0.05, *** p < 0.001 versus unmodified mRNA.
Figure 7.
Nuclease stability of mRNA. mRNA-encoding GLuc in a naked form was incubated in 0.05% serum at 37 °C for 15 min. qRT-PCR measurement was performed to quantify the amount of mRNA that has an intact sequence between a chosen primer pair. The amount of mRNA after serum incubation was compared to that before serum incubation and the relative values are shown in the graph. Data are presented as the mean ± standard error of the mean (S.E.M.) (N = 4). Statistical significance was assessed by one-way analysis of variance (ANOVA) followed by Dunnett’s test. * p < 0.05, *** p < 0.001 versus unmodified mRNA.