Investigating the Protective Effects of a Citrus Flavonoid on the Retardation Morphogenesis of the Oligodendroglia-like Cell Line by Rnd2 Knockdown
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Antibodies and Other Materials
2.3. Synthetic Small Interfering (si)RNAs, Guide (g)RNA-Encoding DNA Sequences, and DNA Primers
2.4. Reverse Transcription (RT)-Polymerase Chain Reaction (PCR)
2.5. Cell Cultures
2.6. siRNA and Grna Transfection Techniques
2.7. Polyacrylamide Gel Electrophoresis and Immunoblotting Following Immunoprecipitation
2.8. Statistical Analyses
2.9. Ethics Statements
3. Results
3.1. Rnd2 Positively Regulates Oligodendroglial Cell Morphological Differentiation
3.2. Rnd2 Effector Molecules Prag1 and Fyn Positively Regulate Morphological Differentiation
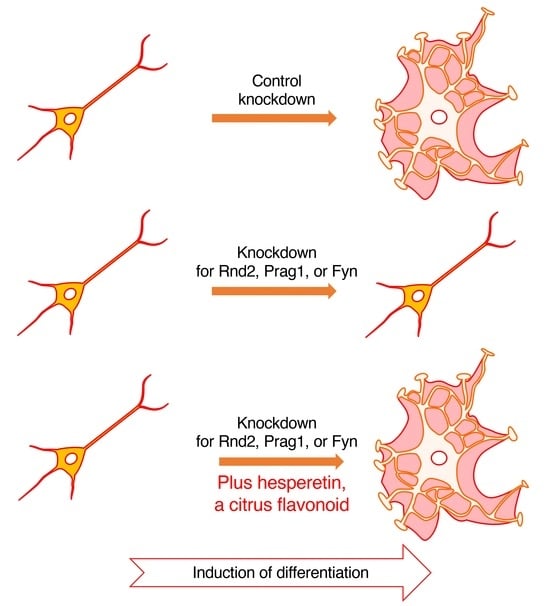
3.3. Hesperetin Recovers Phenotypes Induced by the Knockdown of Rnd2/Prag1/Fyn
3.4. Results Obtained from This Study
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Agarwal, D.; Sandor, C.; Volpato, V.; Caffrey, T.M.; Monzón-Sandoval, J.; Bowden, R.; Alegre-Abarrategui, J.; Wade-Martins, R.; Webber, C. A single-cell atlas of the human substantia nigra reveals cell-specific pathways associated with neurological disorders. Nat. Commun. 2020, 11, 4183. [Google Scholar] [CrossRef] [PubMed]
- Charvériat, M.; Mouthon, F.; Rein, W.; Verkhratsky, A. Connexins as therapeutic targets in neurological and neuropsychiatric disorders. Biochim. Biophys. Acta Mol. Basis Dis. 2021, 1867, 166098. [Google Scholar] [CrossRef] [PubMed]
- Akay, L.A.; Effenberger, A.H.; Tsai, L.H. Cell of all trades: Oligodendrocyte precursor cells in synaptic, vascular, and immune function. Genes Dev. 2021, 35, 180–198. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, N.A.; Fuss, B. Extracellular cues influencing oligodendrocyte differentiation and (re)myelination. Exp. Neurol. 2016, 283 Pt B, 512–530. [Google Scholar] [CrossRef]
- Decourt, B.; Bouleau, Y.; Dulon, D.; Hafidi, A. Expression analysis of neuroleukin, calmodulin, cortactin, and Rho7/Rnd2 in the intact and injured mouse brain. Dev. Brain Res. 2005, 159, 36–54. [Google Scholar] [CrossRef] [PubMed]
- Basbous, S.; Azzarelli, R.; Pacary, E.; Moreau, V. Pathophysiological functions of Rnd proteins. Small GTPases 2021, 12, 336–357. [Google Scholar] [CrossRef] [PubMed]
- Ohtaka-Maruyama, C.; Hirai, S.; Miwa, A.; Heng, J.I.; Shitara, H.; Ishii, R.; Taya, C.; Kawano, H.; Kasai, M.; Nakajima, K.; et al. RP58 regulates the multipolar-bipolar transition of newborn neurons in the developing cerebral cortex. Cell Rep. 2013, 3, 458–471. [Google Scholar] [CrossRef]
- Kerloch, T.; Farrugia, F.; Bouit, L.; Maître, M.; Terral, G.; Koehl, M.; Mortessagne, P.; Heng, J.I.; Blanchard, M.; Doat, H.; et al. The atypical Rho GTPase Rnd2 is critical for dentate granule neuron development and anxiety-like behavior during adult but not neonatal neurogenesis. Mol. Psychiatry 2021, 26, 7280–7295. [Google Scholar] [CrossRef]
- Bar-Sagi, D. A Ras by any other name. Mol. Cell. Biol. 2001, 21, 1441–1443. [Google Scholar] [CrossRef]
- Hall, A. Rho GTPases and the control of cell behaviour. Biochem. Soc. Trans. 2005, 33 Pt 5, 891–895. [Google Scholar] [CrossRef]
- Dahmene, M.; Quirion, L.; Laurin, M. High throughput strategies aimed at closing the GAP in our knowledge of Rho GTPase signaling. Cells 2020, 9, 1430. [Google Scholar] [CrossRef] [PubMed]
- Rossman, K.L.; Der, C.J.; Sondek, J. GEF means go: Turning on RHO GTPases with guanine nucleotide-exchange factors. Nat. Rev. Mol. Cell Biol. 2005, 6, 167–180. [Google Scholar] [CrossRef] [PubMed]
- Riento, K.; Villalonga, P.; Garg, R.; Ridley, A. Function and regulation of RhoE. Biochem. Soc. Trans. 2005, 33 Pt 4, 649–651. [Google Scholar] [CrossRef] [PubMed]
- Chardin, P. Function and regulation of Rnd proteins. Nat. Rev. Mol. Cell Biol. 2006, 7, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, H.; Katoh, H.; Negishi, M. Pragmin, a novel effector of Rnd2 GTPase, stimulates RhoA activity. J. Biol. Chem. 2006, 281, 10355–10364. [Google Scholar] [CrossRef] [PubMed]
- Goh, L.L.; Manser, E. The GTPase-deficient Rnd proteins are stabilized by their effectors. J. Biol. Chem. 2012, 287, 31311–31320. [Google Scholar] [CrossRef] [PubMed]
- Bergles, D.E.; Richardson, W.D. Oligodendrocyte development and plasticity. Cold Spring Harb. Perspect. Biol. 2015, 8, a020453. [Google Scholar] [CrossRef]
- Simons, M.; Nave, K.A. Oligodendrocytes: Myelination and axonal support. Cold Spring Harb. Perspect. Biol. 2015, 8, a020479. [Google Scholar] [CrossRef]
- Tiane, A.; Schepers, M.; Rombaut, B.; Hupperts, R.; Prickaerts, J.; Hellings, N.; van den Hove, D.; Vanmierlo, T. From OPC to oligodendrocyte: An epigenetic journey. Cells 2019, 8, 1236. [Google Scholar] [CrossRef]
- Kuhn, S.; Gritti, L.; Crooks, D.; Dombrowski, Y. Oligodendrocytes in development, myelin generation and beyond. Cells 2019, 8, 1424. [Google Scholar] [CrossRef]
- Barateiro, A.; Brites, D.; Fernandes, A. Oligodendrocyte development and myelination in neurodevelopment: Molecular mechanisms in health and disease. Curr. Pharm. Des. 2016, 22, 656–769. [Google Scholar] [CrossRef] [PubMed]
- Dulamea, A.O. Role of oligodendrocyte dysfunction in demyelination, remyelination and neurodegeneration in multiple sclerosis. Adv. Exp. Med. Biol. 2017, 958, 91–127. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Zhu, Z.; Ransom, B.R.; Tong, X. Oligodendrocyte lineage cells and depression. Mol. Psychiatry 2021, 26, 103–117. [Google Scholar] [CrossRef] [PubMed]
- Clayton, B.L.L.; Tesar, P.J. Oligodendrocyte progenitor cell fate and function in development and disease. Curr. Opin. Cell Biol. 2021, 73, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, Y.; Torii, T.; Terao, M.; Takada, S.; Tanoue, A.; Katoh, H.; Yamauchi, J. Rnd2 differentially regulates oligodendrocyte myelination at different developmental periods. Mol. Biol. Cell 2021, 32, 769–787. [Google Scholar] [CrossRef]
- Horiuchi, M.; Tomooka, Y. An oligodendroglial progenitor cell line FBD-102b possibly secretes a radial glia-inducing factor. Neurosci. Res. 2006, 56, 213–219. [Google Scholar] [CrossRef]
- Okada, A.; Tomooka, Y. A role of Sema6A expressed in oligodendrocyte precursor cells. Neurosci. Lett. 2013, 539, 48–53. [Google Scholar] [CrossRef]
- De Vries, G.H.; Boullerne, A.I. Glial cell lines: An overview. Neurochem. Res. 2010, 35, 1978–2000. [Google Scholar] [CrossRef]
- Tontsch, U.; Archer, D.R.; Dubois-Dalcq, M.; Duncan, I.D. Transplantation of an oligodendrocyte cell line leading to extensive myelination. Proc. Natl. Acad. Sci. USA 1994, 91, 11616–11620. [Google Scholar] [CrossRef]
- Sato, N.; Seiwa, C.; Uruse, M.; Yamamoto, M.; Tanaka, K.; Kawakita, T.; Komatsu, Y.; Yasukawa, A.; Takao, M.; Kudo, C.; et al. Administration of chinpi, a component of the herbal medicine ninjin-youei-to, reverses age-induced demyelination. Evid. Based Complement Alternat. Med. 2011, 2011, 617438. [Google Scholar] [CrossRef]
- Kato, Y.; Tago, K.; Fukatsu, S.; Okabe, M.; Shirai, R.; Oizumi, H.; Ohbuchi, K.; Yamamoto, M.; Mizoguchi, K.; Miyamoto, Y.; et al. CRISPR/CasRx-mediated RNA knockdown reveals that ACE2 is involved in the regulation of oligodendroglial cell morphological differentiation. Noncoding RNA 2022, 8, 42. [Google Scholar] [CrossRef]
- Tago, K.; Kaziro, Y.; Satoh, T. Functional involvement of mSos in interleukin-3 and thrombin stimulation of the Ras, mitogen-activated protein kinase pathway in BaF3 murine hematopoietic cells. J. Biochem. 1998, 123, 659–667. [Google Scholar] [CrossRef] [PubMed]
- Konermann, S.; Lotfy, P.; Brideau, N.J.; Oki, J.; Shokhirev, M.N.; Hsu, P.D. Transcriptome engineering with RNA-targeting type VI-D CRISPR effectors. Cell 2018, 173, 665–676. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.X.; Chong, S.; Zhang, H.; Makarova, K.S.; Koonin, E.V.; Cheng, D.R.; Scott, D.A. Cas13d Is a compact RNA-targeting type VI CRISPR effector positively modulated by a WYL-domain-containing accessory protein. Mol. Cell 2018, 70, 327–339. [Google Scholar] [CrossRef] [PubMed]
- Safari, F.; Murata-Kamiya, N.; Saito, Y.; Hatakeyama, M. Mammalian pragmin regulates Src family kinases via the Glu-Pro-Ile-Tyr-Ala (EPIYA) motif that is exploited by bacterial effectors. Proc. Natl. Acad. Sci. USA 2011, 108, 14938–14943. [Google Scholar] [CrossRef] [PubMed]
- Senda, Y.; Murata-Kamiya, N.; Hatakeyama, M. C-terminal Src kinase-mediated EPIYA phosphorylation of Pragmin creates a feed-forward C-terminal Src kinase activation loop that promotes cell motility. Cancer Sci. 2016, 107, 972–980. [Google Scholar] [CrossRef]
- Umemori, H.; Sato, S.; Yagi, T.; Aizawa, S.; Yamamoto, T. Initial events of myelination involve Fyn tyrosine kinase signalling. Nature 1994, 367, 572–576. [Google Scholar] [CrossRef]
- Matrone, C.; Petrillo, F.; Nasso, R.; Ferretti, G. Fyn tyrosine kinase as harmonizing factor in neuronal functions and dysfunctions. Int. J. Mol. Sci. 2020, 21, 4444. [Google Scholar] [CrossRef]
- Roskoski, R., Jr. Src protein-tyrosine kinase structure and regulation. Biochem. Biophys. Res. Commun. 2004, 324, 1155–1164. [Google Scholar] [CrossRef]
- Okada, M. Regulation of the SRC family kinases by Csk. Int. J. Biol. Sci. 2012, 8, 1385–1397. [Google Scholar] [CrossRef]
- Roskoski, R., Jr. Src protein-tyrosine kinase structure, mechanism, and small molecule inhibitors. Pharmacol. Res. 2015, 94, 9–25. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Wang, H.; Ranjan, K.; Zhang, D. Regulation, targets and functions of CSK. Front. Cell Dev. Biol. 2023, 11, 1206539. [Google Scholar] [CrossRef] [PubMed]
- Spagnuolo, C.; Moccia, S.; Russo, G.L. Anti-inflammatory effects of flavonoids in neurodegenerative disorders. Eur. J. Med. Chem. 2018, 153, 105–115. [Google Scholar] [CrossRef] [PubMed]
- Bandiwadekar, A.; Jose, J.; Khayatkashani, M.; Habtemariam, S.; Khayat Kashani, H.R.; Nabavi, S.M. Emerging novel approaches for the enhanced delivery of natural products for the management of neurodegenerative diseases. J. Mol. Neurosci. 2022, 72, 653–676. [Google Scholar] [CrossRef] [PubMed]
- Nishino, S.; Fujiki, Y.; Sato, T.; Kato, Y.; Shirai, R.; Oizumi, H.; Yamamoto, M.; Ohbuchi, K.; Miyamoto, Y.; Mizoguchi, K.; et al. Hesperetin, a citrus flavonoid, ameliorates inflammatory cytokine-mediated inhibition of oligodendroglial cell morphological differentiation. Neurol. Int. 2022, 14, 471–487. [Google Scholar] [CrossRef] [PubMed]
- Kato, Y.; Shirai, R.; Ohbuchi, K.; Oizumi, H.; Yamamoto, M.; Miyata, W.; Iguchi, T.; Mimaki, Y.; Miyamoto, Y.; Yamauchi, J. Hesperetin ameliorates inhibition of neuronal and oligodendroglial cell differentiation phenotypes induced by knockdown of Rab2b, an autism spectrum disorder-associated gene product. Neurol. Int. 2023, 15, 371–391. [Google Scholar] [CrossRef]
- Li, D.; Mitsuhashi, S.; Ubukata, M. Protective effects of hesperidin derivatives and their stereoisomers against advanced glycation end-products formation. Pharm. Biol. 2012, 50, 1531–1535. [Google Scholar] [CrossRef]
- Yu, H.; Haskins, J.S.; Su, C.; Allum, A.; Haskins, A.H.; Salinas, V.A.; Sunada, S.; Inoue, T.; Aizawa, Y.; Uesaka, M.; et al. In vitro screening of radioprotective properties in the novel glucosylated flavonoids. Int. J. Mol. Med. 2016, 38, 1525–1530. [Google Scholar] [CrossRef]
- Liang, X.; Draghi, N.A.; Resh, M.D. Signaling from integrins to Fyn to Rho family GTPases regulates morphologic differentiation of oligodendrocytes. J. Neurosci. 2004, 24, 7140–7149. [Google Scholar] [CrossRef]
- Madrigal, M.P.; Ballester-Lurbe, B.; Gómez, O.; Moreno-Bravo, J.A.; Puelles, E.; Jurado, S.; Garcia-Verdugo, J.M.; Pérez-Roger, I.; Terrado, J. Rnd3 is necessary for the correct oligodendrocyte differentiation and myelination in the central nervous system. Brain Struct. Funct. 2022, 227, 829–841. [Google Scholar] [CrossRef]
- Heng, J.I.; Nguyen, L.; Castro, D.S.; Zimmer, C.; Wildner, H.; Armant, O.; Skowronska-Krawczyk, D.; Bedogni, F.; Matter, J.M.; Hevner, R.; et al. Neurogenin 2 controls cortical neuron migration through regulation of Rnd2. Nature 2008, 455, 114–118. [Google Scholar] [CrossRef] [PubMed]
- Armentano, M.; Filosa, A.; Andolfi, G.; Studer, M. COUP-TFI is required for the formation of commissural projections in the forebrain by regulating axonal growth. Development 2006, 133, 4151–4162. [Google Scholar] [CrossRef] [PubMed]
- Demuro, S.; Di Martino, R.M.C.; Ortega, J.A.; Cavalli, A. GSK-3b, FYN, and DYRK1A: Master regulators in neurodegenerative pathways. Int. J. Mol. Sci. 2021, 22, 9098. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.; Fu, Y. FYN: Emerging biological roles and potential therapeutic targets in cancer. J. Transl. Med. 2023, 21, 84. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.Y.; Jannat, S.; Jung, H.A.; Choi, J.S. Structural bases for hesperetin derivatives: Inhibition of protein tyrosine phosphatase 1B, kinetics mechanism and molecular molecules. Molecules 2021, 26, 7433. [Google Scholar] [CrossRef]
- Figlia, G.; Gerber, D.; Suter, U. Myelination and mTOR. Glia 2018, 66, 693–707. [Google Scholar] [CrossRef]
- Ogata, T.; Iijima, S.; Hoshikawa, S.; Miura, T.; Yamamoto, S.; Oda, H.; Nakamura, K.; Tanaka, S. Opposing extracellular signal-regulated kinase and Akt pathways control Schwann cell myelination. J. Neurosci. 2004, 24, 6724–6732. [Google Scholar] [CrossRef]
- Dahl, K.D.; Almeida, A.R.; Hathaway, H.A.; Bourne, J.; Brown, T.L.; Finseth, L.T.; Wood, T.L.; Macklin, W.B. mTORC2 loss in oligodendrocyte progenitor cells results in regional hypomyelination in the central nervous system. J. Neurosci. 2023, 43, 540–558. [Google Scholar] [CrossRef]
- Lu, Q.; Lai, Y.; Zhang, H.; Ren, K.; Liu, W.; An, Y.; Yao, J.; Fan, H. Hesperetin inhibits TGF-β1-induced migration and invasion of triple negative breast cancer MDA-MB-231 cells via suppressing Fyn/Paxillin/RhoA pathway. Integr. Cancer Ther. 2022, 21, 15347354221086900. [Google Scholar] [CrossRef]
- Lu, Q.; Kishi, H.; Zhang, Y.; Morita, T.; Kobayashi, S. Hesperetin inhibits sphingosylphosphorylcholine-induced vascular smooth muscle contraction by regulating the Fyn/Rho-Kinase pathway. J. Cardiovasc. Pharmacol. 2022, 79, 456–466. [Google Scholar] [CrossRef]
- Pepper, R.E.; Pitman, K.A.; Cullen, C.L.; Young, K.M. How do cells of the oligodendrocyte lineage affect neuronal circuits to influence motor function, memory and mood? Front. Cell. Neurosci. 2018, 12, 399. [Google Scholar] [CrossRef] [PubMed]
- Sen, M.K.; Mahns, D.A.; Coorssen, J.R.; Shortland, P.J. Behavioural phenotypes in the cuprizone model of central nervous system demyelination. Neurosci. Biobehav. Rev. 2019, 107, 23–46. [Google Scholar] [CrossRef] [PubMed]
- Wegener, A.J.; Neigh, G.N. Animal models of anxiety and depression: Incorporating the underlying mechanisms of sex differences in macroglia biology. Front. Behav. Neurosci. 2021, 15, 780190. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Hong, H.; Liu, C.; Zhang, Y.Q. Social isolation and the brain: Effects and mechanisms. Mol. Psychiatry 2023, 28, 191–201. [Google Scholar] [CrossRef]

















| Reagents or Sources | Company or Source | Cat. No. | Lot. No. | Concentration Used |
|---|---|---|---|---|
| Antibodies | ||||
| Anti-proteolipid protein 1 (PLP1) | Atlas Antibodies | HPA004128 | 8115828 | Immunoblotting (IB), 1:500 |
| Anti-2′,3′-cyclic nucleotides to 2′-nucleotides (CNPase) | Santa Cruz Biotechnology | sc-166559 | A1514 | IB, 1:500 |
| Anti-Sox10 | Santa Cruz Biotechnology | sc-365692 | F1621 | IB, 1:500 |
| Anti-actin (also called pan-b type actin) | MBL | M177-3 | 007 | IB, 1:5000 |
| anti-Fyn | Atlas Antibodies | HPA023887 | A75443 | IB, 1:500; immunopresipitation (IP), 0.1 mg for 300 mg proteins of the respective cell lysates |
| anti-p-c-Src (9A6), which corresponds to anti-(Py420)Fyn | Santa Cruz Biotechnology | sc-81521 | F2122 | IB, 1:100 |
| anti-p-c-Src (H3), which corresponds to anti-(Py531)Fyn | Santa Cruz Biotechnology | sc-166860 | I2921 | IB, 1:100 |
| Anti-IgG (H+L chain) (Rabbit) pAb-HRP | MBL | 458 | 353 | IB, 1:5000 |
| Anti-IgG (H+L chain) (Mouse) pAb-HRP | MBL | 330 | 365 | IB, 1:5000 |
| Key chemicals | ||||
| Hesperetin (Hes) | FUJIFILM Wako Pure Chemical Corporation | 087-10001 | DLK5755 | 15 Mm (final concentration) |
| Monoglucosyl hesperidine (M.Hes) | FUJIFILM Wako Pure Chemical Corporation-Hayashibara Co., Ltd. | 638-07361 | 191127 | 25 Mm (final concentration) |
| Dimethyl sulfoxide (DMSO) | FUJIFILM Wako Pure Chemical Corporation | 047-29353 | CDN0170 | Less than 0.1% (final concentration) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fukatsu, S.; Miyamoto, Y.; Oka, Y.; Ishibashi, M.; Shirai, R.; Ishida, Y.; Endo, S.; Katoh, H.; Yamauchi, J. Investigating the Protective Effects of a Citrus Flavonoid on the Retardation Morphogenesis of the Oligodendroglia-like Cell Line by Rnd2 Knockdown. Neurol. Int. 2024, 16, 33-61. https://doi.org/10.3390/neurolint16010003
Fukatsu S, Miyamoto Y, Oka Y, Ishibashi M, Shirai R, Ishida Y, Endo S, Katoh H, Yamauchi J. Investigating the Protective Effects of a Citrus Flavonoid on the Retardation Morphogenesis of the Oligodendroglia-like Cell Line by Rnd2 Knockdown. Neurology International. 2024; 16(1):33-61. https://doi.org/10.3390/neurolint16010003
Chicago/Turabian StyleFukatsu, Shoya, Yuki Miyamoto, Yu Oka, Maki Ishibashi, Remina Shirai, Yuki Ishida, Shin Endo, Hironori Katoh, and Junji Yamauchi. 2024. "Investigating the Protective Effects of a Citrus Flavonoid on the Retardation Morphogenesis of the Oligodendroglia-like Cell Line by Rnd2 Knockdown" Neurology International 16, no. 1: 33-61. https://doi.org/10.3390/neurolint16010003
APA StyleFukatsu, S., Miyamoto, Y., Oka, Y., Ishibashi, M., Shirai, R., Ishida, Y., Endo, S., Katoh, H., & Yamauchi, J. (2024). Investigating the Protective Effects of a Citrus Flavonoid on the Retardation Morphogenesis of the Oligodendroglia-like Cell Line by Rnd2 Knockdown. Neurology International, 16(1), 33-61. https://doi.org/10.3390/neurolint16010003