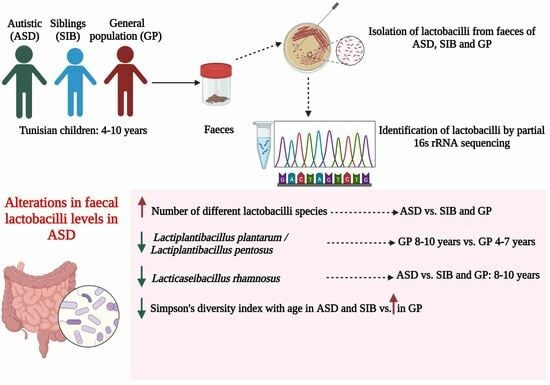
Lactobacilli Profile in Faecal Samples of Tunisian Children Diagnosed with Autism Spectrum Disorder
Abstract
:1. Introduction
2. Materials and Methods
2.1. Subjects and Faecal Sample Collection
2.2. Isolation of Lactobacilli Strains and Culture Conditions
2.3. Identification of Lactobacilli Strains Using Partial 16S rRNA Gene Sequencing
2.4. Prevalence and Species Diversity
2.5. Statistical Analysis
3. Results
3.1. Prevalence, Species Distribution and Characteristics of Lactobacilli in Stools of ASD, SIB and GP Children
3.2. Prevalence, Species Abundance and Diversity of Lactobacilli by Age
3.3. Prevalence, Species Abundance and Diversity of Lactobacilli by Severity of the Disorder
4. Discussion
4.1. Prevalence and Species Abundance of Lactobacilli in Stools of ASD, SIB and GP Groups
4.2. Prevalence, Species Abundance and Diversity of Lactobacilli by Age and Disorder Severity
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Genovese, A.; Butler, M.G. Clinical assessment, genetics, and treatment approaches in autism spectrum disorder (ASD). Int. J. Mol. Sci. 2020, 21, 4726. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Liu, K.; Liu, J.; Liu, Y.W.; Xu, L.; Wang, H.; Zhu, Y.; Wang, P.; Li, Z.; Wen, J.; et al. Dysbiotic Gut Microbiota and Dysregulation of Cytokine Profile in Children and Teens with Autism Spectrum Disorder. Front. Neurosci. 2021, 15, 635925. [Google Scholar] [CrossRef] [PubMed]
- Adams, J.B.; Johansen, L.J.; Powell, L.D.; Quig, D.; Rubin, R.A. Gastrointestinal flora and gastrointestinal status in children with autism—Comparisons to typical children and correlation with autism severity. BMC Gastroenterol. 2011, 11, 22. [Google Scholar] [CrossRef] [PubMed]
- De Angelis, M.; Piccolo, M.; Vannini, L.; Siragusa, S.; De Giacomo, A.; Serrazzanetti, D.I.; Cristofori, F.; Guerzoni, M.E.; Gobbetti, M.; Francavilla, R. Fecal Microbiota and Metabolome of Children with Autism and Pervasive Developmental Disorder Not Otherwise Specified. PLoS ONE 2013, 8, e76993. [Google Scholar] [CrossRef]
- Liu, S.; Li, E.; Sun, Z.; Fu, D.; Duan, G.; Jiang, M.; Yu, Y.; Mei, L.; Yang, P.; Tang, Y.; et al. Altered gut microbiota and short chain fatty acids in Chinese children with autism spectrum disorder. Sci. Rep. 2019, 9, 287. [Google Scholar] [CrossRef]
- Tomova, A.; Husarova, V.; Lakatosova, S.; Bakos, J.; Vlkova, B.; Babinska, K.; Ostatnikova, D. Gastrointestinal microbiota in children with autism in Slovakia. Physiol. Behav. 2015, 138, 179–187. [Google Scholar] [CrossRef]
- Strati, F.; Cavalieri, D.; Albanese, D.; De Felice, C.; Donati, C.; Hayek, J.; Jousson, O.; Leoncini, S.; Renzi, D.; Calabrò, A.; et al. New evidences on the altered gut microbiota in autism spectrum disorders. Microbiome 2017, 5, 24. [Google Scholar] [CrossRef]
- Finegold, S.M.; Dowd, S.E.; Gontcharova, V.; Liu, C.; Henley, K.E.; Wolcott, R.D.; Youn, E.; Summanen, P.H.; Granpeesheh, D.; Dixon, D.; et al. Pyrosequencing study of fecal microflora of autistic and control children. Anaerobe 2010, 16, 444–453. [Google Scholar] [CrossRef]
- Wang, L.; Christophersen, C.T.; Sorich, M.J.; Gerber, J.P.; Angley, M.T.; Conlon, M.A. Low relative abundances of the mucolytic bacterium Akkermansia muciniphila and Bifidobacterium spp. in feces of children with autism. Appl. Environ. Microbiol. 2011, 77, 6718–6721. [Google Scholar] [CrossRef]
- Kang, D.W.; Park, J.G.; Ilhan, Z.E.; Wallstrom, G.; LaBaer, J.; Adams, J.B.; Krajmalnik-Brown, R. Reduced Incidence of Prevotella and Other Fermenters in Intestinal Microflora of Autistic Children. PLoS ONE 2013, 8, e68322. [Google Scholar] [CrossRef]
- Pulikkan, J.; Maji, A.; Dhakan, D.B.; Saxena, R.; Mohan, B.; Anto, M.M.; Agarwal, N.; Grace, T.; Sharma, V.K. Gut Microbial Dysbiosis in Indian Children with Autism Spectrum Disorders. Microb. Ecol. 2018, 76, 1102–1114. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.M.A.P.; Harris, H.M.B.; Mattarelli, P.; O’toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef] [PubMed]
- Gänzle, M.G. Lactic metabolism revisited: Metabolism of lactic acid bacteria in food fermentations and food spoilage. Curr. Opin. Food Sci. 2015, 2, 106–117. [Google Scholar] [CrossRef]
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders, 5th ed.; DSM-5; American Psychiatric Association: Washington, DC, USA, 2013. [Google Scholar] [CrossRef]
- Lord, C.; Rutter, M.; Le Couteur, A. Autism Diagnostic Interview-Revised: A Revised Version of a Diagnostic Interview for Caregivers of Individuals with Possible Pervasive Developmental Disorders. J. Autism Dev. Disord. 1994, 24, 659–685. [Google Scholar] [CrossRef]
- Lord, C.; Rutter, M.; Goode, S.; Heemsbergen, J.; Jordan, H.; Mawhood, L.; Schopler, E. Autism diagnostic observation schedule: A standardized observation of communicative and social behavior. J. Autism Dev. Disord. 1989, 19, 185–212. [Google Scholar] [CrossRef]
- Schopler, E.; Reichler, R.J.; DeVellis, R.F.; Daly, K. Toward objective classification of childhood autism: Childhood Autism Rating Scale (CARS). J. Autism Dev. Disord. 1980, 10, 91–103. [Google Scholar] [CrossRef]
- Chamtouri, M.; Merghni, A.; Salazar, N.; Redruello, B.; Gaddour, N.; Mastouri, M.; Arboleya, S.; de Los Reyes-Gavilán, C.G. An Overview on Fecal Profiles of Amino Acids and Related Amino-Derived Compounds in Children with Autism Spectrum Disorder in Tunisia. Molecules 2023, 28, 3269. [Google Scholar] [CrossRef]
- Derriche, I.; Nogacka, A.M.; Salazar, N.; Ruas-Madiedo, P.; Gueimonde, M.; Bensalah, F.; de Los Reyes-Gavilán, C.G. Effect of inulin-type fructans and galactooligosaccharides on cultures of Lactobacillus strains isolated in Algeria from camel’s milk and human colostrum. Food Sci. Technol. Int. 2021, 27, 223–233. [Google Scholar] [CrossRef]
- Kullen, M.J.; Sanozky-Dawes, R.B.; Crowell, D.C.; Klaenhammer, T.R. Use of the DNA sequence of variable regions of the 16S rRNA gene for rapid and accurate identification of bacteria in the Lactobacillus acidophilus complex. J. Appl. Microbiol. 2000, 89, 511–516. [Google Scholar] [CrossRef]
- Simpson, E. Measurement of Diversity. Nature 1949, 163, 688. [Google Scholar] [CrossRef]
- Kim, B.R.; Shin, J.; Guevarra, R.B.; Lee, J.H.; Kim, D.W.; Seol, K.H.; Lee, J.H.; Kim, H.B.; Isaacson, R. Deciphering diversity indices for a better understanding of microbial communities. J. Microbiol. Biotechnol. 2017, 27, 2089–2093. [Google Scholar] [CrossRef]
- Ortiz-Burgos, S. Shannon-Weaver Diversity Index. In Encyclopedia of Estuaries, 1st ed.; Kennish, M.J., Ed.; Springer: Dordrecht, The Netherlands, 2016; pp. 572–573. [Google Scholar] [CrossRef]
- O’Hara, A.M.; O’Regan, P.; Fanning, Á.; O’Mahony, C.; MacSharry, J.; Lyons, A.; Bienenstock, J.; O’Mahony, L.; Shanahan, F. Functional modulation of human intestinal epithelial cell responses by Bifidobacterium infantis and Lactobacillus salivarius. Immunology 2006, 118, 202–215. [Google Scholar] [CrossRef] [PubMed]
- Ashwood, P.; Krakowiak, P.; Hertz-Picciotto, I.; Hansen, R.; Pessah, I.; Van de Water, J. Elevated plasma cytokines in autism spectrum disorders provide evidence of immune dysfunction and are associated with impaired behavioral outcome. Brain Behav. Immun. 2011, 25, 40–45. [Google Scholar] [CrossRef] [PubMed]
- Yun, S.W.; Kim, J.K.; Lee, K.E.; Oh, Y.J.; Choi, H.J.; Han, M.J.; Kim, D.H. A probiotic Lactobacillus gasseri alleviates Escherichia coli-induced cognitive impairment and depression in mice by regulating il-1β expression and gut microbiota. Nutrients 2020, 12, 3441. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.K.; Lee, K.E.; Lee, S.A.; Jang, H.M.; Kim, D.H. Interplay Between Human Gut Bacteria Escherichia coli and Lactobacillus mucosae in the Occurrence of Neuropsychiatric Disorders in Mice. Front. Immunol. 2020, 1, 273. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, T.S.; Arnoux, J.; O’Toole, P.W. Metagenomic analysis reveals distinct patterns of gut Lactobacillus prevalence, abundance, and geographical variation in health and disease. Gut Microbes 2020, 12, 1822729. [Google Scholar] [CrossRef] [PubMed]
- Mathipa-Mdakane, M.G.; Thantsha, M.S. Lacticaseibacillus rhamnosus: A Suitable Candidate for the Construction of Novel Bioengineered Probiotic Strains for Targeted Pathogen Control. Foods 2022, 11, 785. [Google Scholar] [CrossRef]
- Johansson, M.A.; Björkander, S.; Mata Forsberg, M.; Qazi, K.R.; Salvany Celades, M.; Bittmann, J.; Eberl, M.; Sverremark-Ekström, E. Probiotic lactobacilli modulate Staphylococcus aureus-induced activation of conventional and unconventional T cells and NK cells. Front. Immunol. 2016, 7, 273. [Google Scholar] [CrossRef]
- Chen, J.F.; Zhuang, Y.; Jin, S.B.; Zhang, S.L.; Yang, W.W. Probiotic Lactobacillus rhamnosus GG (LGG) restores intestinal dysbacteriosis to alleviate upregulated inflammatory cytokines triggered by femoral diaphyseal fracture in adolescent rodent model. Eur. Rev. Med. Pharmacol. Sci. 2021, 25, 376–389. [Google Scholar] [CrossRef]
- Drago, L.; Nicola, L.; Iemoli, E.; Banfi, G.; De Vecchi, E. Strain-dependent release of cytokines modulated by Lactobacillus salivarius human isolates in an in vitro model. BMC Res. Notes 2010, 3, 44. [Google Scholar] [CrossRef]
- Yao, Y.; Cai, X.; Ye, Y.; Wang, F.; Chen, F.; Zheng, C. The Role of Microbiota in Infant Health: From Early Life to Adulthood. Front. Immunol. 2021, 12, 708472. [Google Scholar] [CrossRef] [PubMed]


| Lactobacilli Species by Fermentation Type | |||
|---|---|---|---|
| Homofermentative n (%) | Heterofermentative n (%) | ||
| Whole three groups | ASD | 19 (90.5) | 2 (9.5) |
| SIB | 12 (100) | 0 (0) | |
| GP | 15 (88.2) | 2 (11.8) | |
| p-value | 0.487 | ||
| 4–7 years | ASD 4–7 | 7 (100) | 0 (0) |
| SIB 4–7 | 7 (100) | 0 (0) | |
| GP 4–7 | 10 (100) | 0 (0) | |
| p-value | - | ||
| 8–10 years | ASD 8–10 | 12 (85.7) | 2 (14.3) |
| SIB 8–10 | 5 (100) | 0 (0) | |
| GP 8–10 | 5 (71.4) | 2 (28.6) | |
| p-value | 0.395 | ||
| ASD severity | Mild-to-moderate ASD | 5 (83.3) | 1 (16.7) |
| Severe ASD | 14 (93.3) | 1 (6.7) | |
| p-value | 0.5 | ||
| Simpson’s Diversity Index | Shannon–Wiener Diversity Index | ||||
|---|---|---|---|---|---|
| All Species | Homofermentative Species | All Species | Homofermentative Species | ||
| Whole three groups | ASD | 0.8 | 0.8 | 1.7 | 1.5 |
| SIB | 0.8 | 0.8 | 1.4 | 1.4 | |
| GP | 0.8 | 0.7 | 1.5 | 1.2 | |
| 4–7 years | ASD 4–7 | 0.8 | 0.8 | 1.3 | 1.3 |
| SIB 4–7 | 0.9 | 0.9 | 1.5 | 1.5 | |
| GP 4–7 | 0.6 | 0.6 | 0.9 | 0.9 | |
| 8–10 years | ASD 8–10 | 0.8 | 0.7 | 1.6 | 1.2 |
| SIB 8–10 | 0.7 | 0.7 | 0.7 | 0.7 | |
| GP 8–10 | 0.9 | 0.8 | 1.4 | 1.1 | |
| ASD severity | Mild-to-moderate ASD | 0.8 | 0.7 | 1.2 | 1 |
| Severe ASD | 0.8 | 0.8 | 1.6 | 1.4 | |
| Abundance | Number of Lactobacilli Isolates from ASD (n = 21) | Number of Lactobacilli Isolates from SIB (n = 12) | Number of Lactobacilli Isolates from GP (n = 17) | ||||
|---|---|---|---|---|---|---|---|
| 4–7 y (n = 7) | 8–10 y (n = 14) | 4–7 y (n = 7) | 8–10 y (n = 5) | 4–7 y (n = 10) | 8–10 y (n = 7) | ||
| Ligilactobacillus salivarius | Number of isolates, n (%) | 1 (14.3) | 7 (50) | 1 (14.3) | 0 (0) | 4 (40) | 2 (28.6) |
| p-value | 0.17 | 1 | 1 | ||||
| Lacticaseibacillus rhamnosus | Number of isolates, n (%) | 3 (42.9) | 1 (7.1) | 1 (14.3) | 3 (60) | 1 (10) | 2 (28.6) |
| p-value | 0.09 | 0.22 | 0.54 | ||||
| Lactobacillus gasseri | Number of isolates, n (%) | 2 (28.6) | 1 (7.1) | 1 (14.3) | 0 (0) | - | - |
| p-value | 0.25 | 1 | |||||
| Lactiplantibacillus plantarum/Lactiplantibacillus pentosus | Number of isolates, n (%) | 1 (14.3) | 1 (7.1) | 3 (42.9) | 2 (40) | 5 (50) | 0 (0) |
| p-value | 1 | 1 | 0.04 * | ||||
| Lacticaseibacillus paracasei/Lacticaseibacillus casei | Number of isolates, n (%) | 0 (0) | 2 (14.3) | 1 (14.3) | 0 (0) | 0 (0) | 1 (14.3) |
| p-value | 0.53 | 1 | 0.41 | ||||
| Limosilactobacillus mucosae | Number of isolates, n (%) | 0 (0) | 1 (7.1) | - | - | - | - |
| p-value | 1 | ||||||
| Limosilactobacillus fermentum | Number of isolates, n (%) | 0 (0) | 1 (7.1) | - | - | 0 (0) | 2 (28.6) |
| p-value | 1 | 0.15 | |||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chamtouri, M.; Merghni, A.; Gaddour, N.; Mastouri, M.; Arboleya, S.; de los Reyes-Gavilán, C.G. Lactobacilli Profile in Faecal Samples of Tunisian Children Diagnosed with Autism Spectrum Disorder. Microbiol. Res. 2023, 14, 1225-1237. https://doi.org/10.3390/microbiolres14030082
Chamtouri M, Merghni A, Gaddour N, Mastouri M, Arboleya S, de los Reyes-Gavilán CG. Lactobacilli Profile in Faecal Samples of Tunisian Children Diagnosed with Autism Spectrum Disorder. Microbiology Research. 2023; 14(3):1225-1237. https://doi.org/10.3390/microbiolres14030082
Chicago/Turabian StyleChamtouri, Mariem, Abderrahmen Merghni, Naoufel Gaddour, Maha Mastouri, Silvia Arboleya, and Clara G. de los Reyes-Gavilán. 2023. "Lactobacilli Profile in Faecal Samples of Tunisian Children Diagnosed with Autism Spectrum Disorder" Microbiology Research 14, no. 3: 1225-1237. https://doi.org/10.3390/microbiolres14030082
APA StyleChamtouri, M., Merghni, A., Gaddour, N., Mastouri, M., Arboleya, S., & de los Reyes-Gavilán, C. G. (2023). Lactobacilli Profile in Faecal Samples of Tunisian Children Diagnosed with Autism Spectrum Disorder. Microbiology Research, 14(3), 1225-1237. https://doi.org/10.3390/microbiolres14030082