Sustainability of Water Reclamation: Long-Term Recharge with Reclaimed Wastewater Does Not Enhance Antibiotic Resistance in Sediment Bacteria
Abstract
:1. Introduction
2. Materials and Methods
2.1. Field Sampling

2.2. Laboratory Isolation and Confirmation of Enterococcus
2.3. Screening of Isolates for Antibiotic Sensitivity
| Antibiotic | Abbreviation | Antibiotic Class | Concentration Range (µg·mL−1) | Resistance Breakpoint (µg·mL−1) |
|---|---|---|---|---|
| Kanamycin | KAN | Aminoglycoside | 128–1024 | ≥1024 |
| Linezolid | LZD | Oxazoladinone | 0.5–8 | ≥8 |
| Lincomycin | LIN | Lincosamide | 1–8 | ≥16 |
| Quinupristin/dalfopristin | SYN | Streptogramin | 0.5–32 | ≥4 |
| Gentamicin | GEN | Aminoglycoside | 128–1024 | >500 |
| Tylosin tartrate | TYLT | Macrolide | 0.25–32 | ≥32 |
| Vancomycin | VAN | Glycopeptide | 0.25–32 | ≥32 |
| Daptomycin | DAP | Other | 0.25–16 | ≥8 |
| Penicillin | PEN | Β-lactam | 0.25–16 | ≥16 |
| Chloramphenicol | CHL | Other | 2–32 | ≥32 |
| Ciprofloxacin | CIP | Fluoroquinolone | 0.12–4 | ≥4 |
| Tetracycline | TET | Tetracycline | 1–32 | ≥16 |
| Erythromycin | ERY | Macrolide | 0.25–8 | ≥8 |
| Tigecycline | TIG | Glycylcycline | 0.015–0.5 | >0.5 a |
| Streptomycin | STR | Aminoglycoside | 512–2048 | >1024 |
| Nitrofurantoin | NIT | Other | 2–64 | ≥128 |
2.4. Statistical Analysis
3. Results
3.1. Antibiotic Resistance
| Antibiotic | Treated Wastewater Sites | Groundwater Sites | ||||
|---|---|---|---|---|---|---|
| Total # Isolates | Resistance Rate (%) | MIC50 (µg·mL−1) | Total # Isolates | Resistance Rate (%) | MIC50 (µg·mL−1) | |
| Kanamycin | 264 | 3.8 | <128 | 134 | ND | <128 |
| Linezolid | 264 | 4.5 | 1 | 134 | 4.5 | 0.5 |
| Lincomycin | 264 | 60.6 | 4 | 134 | 70.1 | 8 |
| Quinupristin/dalfopristin | 264 | 15.2 | 0.5 | 134 | 19.4 | 0.5 |
| Gentamicin | 264 | 1.5 | <128 | 134 | ND | <128 |
| Tylosin tartrate | 264 | 2.3 | 0.5 | 134 | 13.4 | 2 |
| Vancomycin | 264 | 6.1 | 2 | 134 | 7.5 | 1 |
| Daptomycin | 264 | 16.7 | 2 | 134 | 19.4 | 1 |
| Penicillin | 264 | 7.6 | 1 | 134 | 9.0 | 1 |
| Chloramphenicol | 264 | 3.0 | 2 | 134 | ND | 4 |
| Ciprofloxacin | 264 | 18.2 | 1 | 134 | 29.9 | 1 |
| Tetracycline | 264 | 3.0 | 1 | 134 | 13.4 | 1 |
| Erythromycin | 264 | 19.7 | 2 | 134 | 34.3 | 4 |
| Tigecycline | 264 | 9.1 | 0.12 | 134 | 13.4 | 0.12 |
| Streptomycin | 264 | ND | <512 | 134 | ND | <512 |
| Nitrofurantoin | 264 | ND | 8 | 134 | ND | 16 |
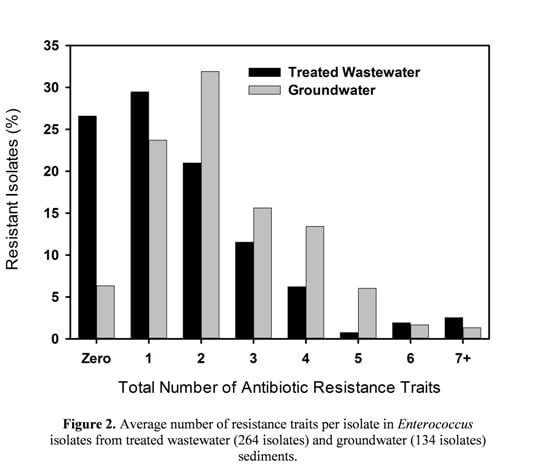
3.2. Multiple Antibiotic Resistance and Resistance Profiles

3.3. MIC50
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- WHO (World Health Organization). WHO Annual Report on Infectious Disease: Overcoming Antimicrobial Resistance. Available online: http://www.who.int/infectious-disease-report/2000/ (accessed on 20 January 2014).
- Böckelmann, U.; Dörries, H.-H.; Ayuso-Gabella, M.N.; de Marçay, M.S.; Tandoi, V.; Levantesi, C.; Masciopinto, C.; van Houtte, E.; Szewzyk, U.; Wintgens, T.; et al. Quantitative PCR monitoring of antibiotic resistance genes and bacterial pathogens in three European artificial groundwater recharge systems. Appl. Environ. Microbiol. 2009, 75, 154–163. [Google Scholar] [CrossRef]
- Miao, X.S.; Bishay, F.; Chen, M.; Matcalfe, C.D. Occurrence of antimicrobials in the final effluents of wastewater treatment plants in Canada. Environ. Sci. Technol. 2004, 38, 3533–3541. [Google Scholar] [CrossRef]
- Göbel, A.; Thomsen, A.; McArdell, C.S.; Joss, A.; Giger, W. Occurrence and sorption behavior of sulfonamides, macrolides, and trimethoprim in activated sludge treatment. Environ. Sci. Technol. 2005, 39, 3981–3989. [Google Scholar] [CrossRef]
- Jury, K.L.; Khan, S.J.; Vancov, T.; Stuetz, R.M.; Ashbolt, N.J. Are sewage treatment plants promoting antibiotic resistance? Crit. Rev. Environ. Sci. Technol. 2011, 41, 243–270. [Google Scholar] [CrossRef]
- Cha, J.M.; Yang, S.; Carlson, K.H. Trace determination of β-lactam antibiotics in surface water and urban wastewater using liquid chromatography combined with electrospray tandem mass spectrometry. J. Chromatogr. A 2006, 1115, 46–57. [Google Scholar]
- Göbel, A.; McArdell, C.S.; Joss, A.; Siegrist, H.; Giger, W. Fate of sulfonamides, macrolides, and trimethoprim in different wastewater treatment technologies. Sci. Total Environ. 2007, 372, 361–371. [Google Scholar] [CrossRef]
- Lindberg, R.H.; Wennberg, P.; Johansson, M.I.; Tysklind, M.; Andersson, B.A.V. Screening of human antibiotic substances and determination of weekly mass flows in five sewage treatment plants in Sweden. Environ. Sci. Technol. 2005, 39, 3421–3429. [Google Scholar] [CrossRef]
- Karthikeyan, K.G.; Meyer, M.T. Occurrence of antibiotics in wastewater treatment facilities in Wisconsin, USA. Sci. Total Environ. 2006, 361, 196–207. [Google Scholar] [CrossRef]
- Kolpin, D.W.; Furlong, E.T.; Meyer, M.T.; Thurman, E.M.; Zaugg, S.D.; Barber, L.B.; Buxton, H.T. Pharmaceuticals, hormones, and other organic wastewater contaminants in US streams, 1999–2000: A national reconnaissance. Environ. Sci. Technol. 2002, 36, 1202–1211. [Google Scholar]
- Yang, S.; Carlson, K.H. Evolution of antibiotic (tetracyclines and sulfonamides) occurrence in a river through pristine, urban and agricultural landscapes. Water Res. 2003, 37, 4645–4656. [Google Scholar] [CrossRef]
- Pauwels, B.; Verstraete, W. The treatment of hospital wastewater: An appraisal. J. Water Health 2006, 4, 405–416. [Google Scholar]
- Auerbach, E.A.; Seyfried, E.E.; McMahon, K.D. Tetracycline resistance genes in activated sludge wastewater treatment plants. Water Res. 2007, 41, 1143–1151. [Google Scholar] [CrossRef]
- Giger, W.; Alder, A.C.; Golet, E.M.; Kohler, H.E.; McArdell, C.S.; Molnar, E.; Siegrist, H.; Suter, M.F. Occurrence and fate of antibiotics as trace contaminants in wastewaters, sewage sludges, and surface waters. Chimia 2003, 57, 485–491. [Google Scholar] [CrossRef]
- Dhanapal, L.P.; Morse, A.N. Effect of analgesics and their derivatives on antibiotic resistance of environmental microbes. Water Sci. Technol. 2009, 59, 1823–1829. [Google Scholar] [CrossRef]
- Waksman, S.A.; Woodruff, H.B. The soil as a source of microorganisms antagonistic to disease-producing bacteria. J. Bacteriol. 1940, 40, 581–600. [Google Scholar]
- Martin, M.F.; Liras, P. Organization and expression of genes involved in the biosynthesis of antibiotics and other secondary metabolites. Annu. Rev. Microbiol. 1989, 43, 173–206. [Google Scholar] [CrossRef]
- Hopwood, D.A. How do antibiotic-producing bacteria ensure their self-resistance before antibiotic biosynthesis incapacitates them? Mol. Microbiol. 2007, 63, 937–940. [Google Scholar] [CrossRef]
- D’Costa, V.M.; McGrann, K.M.; Hughes, D.W.; Wright, G.D. Sampling the antibiotic resistome. Science 2006, 311, 374–377. [Google Scholar] [CrossRef]
- Brown, M.G.; Balkwill, D.L. Antibiotic resistance in bacteria isolated from the deep terrestrial subsurface. Microb. Ecol. 2009, 57, 484–493. [Google Scholar] [CrossRef]
- Hayes, J.R.; English, L.L.; Carr, L.E.; Wagner, D.D.; Joseph, S.W. Multiple-antibiotic resistance of Enterococcus spp. isolated from commercial poultry production environments. Appl. Environ. Microbiol. 2004, 70, 6005–6011. [Google Scholar]
- Giard, J.C.; Laplace, J.M.; Rince, A.; Pichereau, V.; Benachour, A.; Leboeuf, C.; Flahaut, S.; Auffray, Y.; Artke, A. The stress proteome of Enterococcus faecalis. Electrophoresis 2001, 22, 2947–2954. [Google Scholar] [CrossRef]
- Kühn, I.; Iversen, A.; Burman, I.G.; Olsson-Liljequist, B.; Franklin, A.; Finn, M.; Aarestrup, F.; Seyfarth, A.M.; Blanch, A.R.; Taylor, H.; et al. Epidemiology and ecology of enterococci, with special reference to antibiotic resistant strains in animals, humans, and the environment. Example of an ongoing project within the European research programme. Int. J. Antimicrob. Agents 2000, 14, 337–342. [Google Scholar] [CrossRef]
- Kayser, F.H. Safety aspects of enterococci from the medical point of view. Int. J. Food Microbiol. 2003, 88, 255–262. [Google Scholar] [CrossRef]
- EUCAST (European Committee on Antimicrobial Susceptibility Testing) Steering Committee. EUCAST technical note on tigecycline. Clin. Microbiol. Infec. 2006, 12, 1147–1149. [Google Scholar] [CrossRef]
- He, J.-W.; Jiang, S. Quantification of enterococci and human adenovirus in environmental samples by real-time PCR. Appl. Environ. Microbiol. 2005, 71, 2250–2255. [Google Scholar] [CrossRef]
- Post, D.F.; Mack, C.; Camp, P.D.; Suliman, A.S. Mapping and characterization of the soils on the University of Arizona Maricopa Agricultural Center. J. Ariz.-Nev. Acad. Sci. 1988, 18, 49–60. [Google Scholar]
- Harper, W.G.; Youngs, F.O.; Strahorn, A.T.; Armstrong, S.W.; Schwalen, H.C. Soil survey of the Salt River Valley Area, Arizona; U.S. Department of Agriculture Bureau of Chemistry and Soils: Washington, DC, USA, 1926. [Google Scholar]
- CLSI (Clinical and Laboratory Standards Institute). Performance Standards for Antimicrobial Susceptibility Testing; Twentieth Informational Supplement; Document M100-S20; CLSI: Wayne, PA, USA, 2010. [Google Scholar]
- Chapin, K.C.; Musgnug, M.C. Validation of the automated reading and incubation system with Sensititre® plates for antimicrobial susceptibility testing. J. Clin. Microbiol. 2003, 41, 1951–1956. [Google Scholar] [CrossRef]
- Schwarz, S.; Silley, P.; Simjee, S.; Woodfords, N.; van Duijkeren, E.; Johnson, A.P.; Gaastra, W. Assessing the antimicrobial susceptibility of bacteria obtained from animals. J. Antimicrob. Chemoth. 2010, 65, 601–604. [Google Scholar] [CrossRef]
- Kim, S.; Aga, D.S. Potential ecological and human health impacts of antibiotics and antibiotic-resistant bacteria from wastewater treatment plants. J. Toxicol. Environ. Health Part B 2007, 10, 559–573. [Google Scholar] [CrossRef]
- Gullberg, E.; Cao, S.; Berg, O.G.; Ilback, C.; Sandegren, L.; Hughes, D.; Andersson, D.I. Selection of resistant bacteria at very low antibiotic concentrations. PLoS Pathog. 2011, 7, e1002158. [Google Scholar] [CrossRef]
- Silver, S.; Phung, L.T. Bacterial heavy metal resistance: New surprises. Annu. Rev. Microbiol. 1996, 50, 753–789. [Google Scholar] [CrossRef]
- Alonso, A.; Sanchez, P.; Martinez, J.L. Environmental selection of antibiotic resistance genes. Environ. Microbiol. 2001, 3, 1–9. [Google Scholar] [CrossRef]
- Baker-Austin, C.; Wright, M.S.; Stepanauskas, R.; McArthur, J.V. Co-selection of antibiotic and metal resistance. Trends Microbiol. 2006, 14, 176–182. [Google Scholar] [CrossRef]
- Williams, C.F.; McLain, J.E.T. Soil persistence and fate of carbamazepine, lincomycin, caffeine, and ibuprofen from wastewater reuse. J. Environ. Qual. 2012, 41, 1473–1480. [Google Scholar] [CrossRef]
- Brown, K.D.; Kulis, J.; Thomson, B.; Chapman, T.H.; Mawhinney, D.B. Occurrence of antibiotics in hospital, residential, and dairy effluent, municipal wastewater, and the Rio Grande in New Mexico. Sci. Total Environ. 2006, 366, 772–783. [Google Scholar] [CrossRef]
- Diwan, V.; Lundborg, C.S.; Tamhanka, A.J. Seasonal and temporal variation in release of antibiotics in hospital wastewater: Estimation using continuous and grab sampling. PLoS One 2013, 8, 1–7. [Google Scholar]
- Andersen, S.R. Effects of waste water treatment on the species composition and antibiotic resistance of coliform bacteria. Curr. Microbiol. 1993, 26, 97–103. [Google Scholar] [CrossRef]
- Galvin, S.; Boyle, F.; Hickey, P.; Vellinga, A.; Morris, D.; Cormican, M. Enumeration and characterization of antimicrobial-resistant Escherichia coli bacteria in effluent from municipal, hospital, and secondary treatment facility sources. Appl. Environ. Microbiol. 2010, 76, 4772–4779. [Google Scholar] [CrossRef]
- Reinthaler, F.F.; Posch, J.; Feieri, G.; Wust, G.; Haas, D.; Ruckenbauer, G.; Mascher, F.; Marth, E. Antibiotic resistance of E. coli in sewage and sludge. Water Res. 2003, 37, 1685–1690. [Google Scholar] [CrossRef]
- Harwood, V.J.; Whitlock, J.; Withington, V. Classification of antibiotic resistance patterns of indicator bacteria by discriminant analysis use in predicting the source of fecal contamination in subtropical waters. Appl. Environ. Microbiol. 2000, 66, 3698–3704. [Google Scholar] [CrossRef]
- Garcia-Armisen, T.; Vercammen, K.; Passerat, J.; Triest, D.; Servais, P.; Cornelis, P. Antimicrobial resistance of heterotrophic bacteria in sewage-contaminated rivers. Water Res. 2011, 45, 788–796. [Google Scholar] [CrossRef]
- Figueira, V.; Serra, E.; Manaia, C.M. Differential patterns of antimicrobial resistance in population subsets of Escherichia coli isolated from waste- and surface waters. Sci. Total Environ. 2011, 409, 1017–1023. [Google Scholar] [CrossRef]
- Murray, B.E. The life and times of the Enterococcus. Clin. Microbiol. Rev. 1990, 3, 46–65. [Google Scholar]
- Klare, I.; Konstabel, C.; Badstübner, D.; Werner, G.; Witte, W. Occurrence and spread of antibiotic resistances in Enterococcus faecium. Int. J. Food Mirobiol. 2003, 88, 269–290. [Google Scholar] [CrossRef]
- Luczkiewicz, A.; Jankowska, K.; Bray, R.; Kulbat, E.; Quant, B.; Sokolowska, A.; Olanczuk-Neyman, K. Antimicrobial resistance of fecal indicators in disinfected wastewater. Water Sci. Technol. 2011, 64, 2352–2361. [Google Scholar] [CrossRef]
- Servais, P.; Passerat, J. Antimicrobial resistance of fecal bacteria in waters of the Seine river watershed (France). Sci. Total Environ. 2009, 408, 365–372. [Google Scholar] [CrossRef]
- Abriouel, H.; Omar, N.B.; Molinos, A.C.; López, R.L.; Grande, M.J.; Martinez-Viedma, P.; Ortega, E.; Cañamero, M.M.; Galvez, A. Comparative analysis of genetic diversity and incidence of virulence factors and antibiotic resistance among enterococcal populations from raw fruit and vegetable foods, water and soil, and clinical samples. Int. J. Food Microbiol. 2008, 123, 38–49. [Google Scholar] [CrossRef]
- Andrews, J.M. Determination of minimum inhibitory concentrations. J. Antimicrob. Chemother. 2001, 48, 5–16. [Google Scholar] [CrossRef]
- Kohanski, M.A.; DePristo, M.A.; Collins, J.J. Sublethal antibiotic treatment leads to multidrug resistance via radical-induced mutagenesis. Mol. Cell 2010, 37, 311–320. [Google Scholar] [CrossRef]
- Negreanu, Y.; Pasternak, A.; Jurkevitch, E.; Cytryn, E. Impact of treated wastewater irrigation on antibiotic resistance in agricultural soils. Env. Sci. Technol. 2012, 46, 4800–4808. [Google Scholar] [CrossRef]
- Rosenblatt-Farrell, N. The landscape of antibiotic resistance. Environ. Health Perspect. 2009, 117, A244–A250. [Google Scholar] [CrossRef]
- American Academy of Microbiology (AAM). Antibiotic Resistance: An Ecological Perspective to an Old Problem; AAM: Washington, DC, USA, 2009. [Google Scholar]
- Lima-Bittencourt, C.I.; Cursino, L.; Gonçalves-Dornelas, H.; Pontes, D.S.; Nardi, R.M.; Callisto, M.; Chartone-Souza, E.; Nascimento, A.M.A. Multiple antimicrobial resistance in Enterobacteriaceae isolates from pristine freshwater. Genet. Mol. Res. 2007, 6, 510–521. [Google Scholar]
- D’Costa, V.M.; King, C.F.; Kalan, L.; Morar, M.; Sung, W.W.L.; Schwarz, C.; Froese, D.; Zazula, G.; Calmels, F.; Debruyne, R.; et al. Antibiotic resistance is ancient. Nature 2011, 477, 457–461. [Google Scholar] [CrossRef]
- Davies, J. Are antibiotics naturally antibiotics? J. Ind. Microbiol. Biotechnol. 2006, 33, 496–499. [Google Scholar] [CrossRef]
- Fajardo, A.; Martinez, J.L. Antibiotics as signals that trigger specific bacterial responses. Curr. Opin. Microbiol. 2008, 11, 161–167. [Google Scholar] [CrossRef]
- Linares, J.F.; Gustafsson, I.; Baquero, F.; Martinez, J.L. Antibiotics as inter-microbial signaling agents instead of weapons. Proc. Natl. Acad. Sci. USA 2006, 103, 19484–19489. [Google Scholar] [CrossRef]
- Wardle, D.A. A comparative assessment of the factors which influence microbial biomass carbon and nitrogen levels in soil. Biol. Rev. 1992, 67, 321–358. [Google Scholar] [CrossRef]
- Neff, J.C.; Asner, G.P. Dissolved organic carbon in terrestrial ecosystems: Synthesis and a model. Ecosystems 2001, 4, 29–48. [Google Scholar] [CrossRef]
- Martinez, J.L.; Rojo, F. Metabolic regulation of antibiotic resistance. FEMS Microbiol. Rev. 2011, 35, 768–789. [Google Scholar] [CrossRef]
- Amman, R.I.; Ludwig, W.; Schleifer, K.H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 1995, 59, 143–169. [Google Scholar]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
McLain, J.E.; Williams, C.F. Sustainability of Water Reclamation: Long-Term Recharge with Reclaimed Wastewater Does Not Enhance Antibiotic Resistance in Sediment Bacteria. Sustainability 2014, 6, 1313-1327. https://doi.org/10.3390/su6031313
McLain JE, Williams CF. Sustainability of Water Reclamation: Long-Term Recharge with Reclaimed Wastewater Does Not Enhance Antibiotic Resistance in Sediment Bacteria. Sustainability. 2014; 6(3):1313-1327. https://doi.org/10.3390/su6031313
Chicago/Turabian StyleMcLain, Jean E., and Clinton F. Williams. 2014. "Sustainability of Water Reclamation: Long-Term Recharge with Reclaimed Wastewater Does Not Enhance Antibiotic Resistance in Sediment Bacteria" Sustainability 6, no. 3: 1313-1327. https://doi.org/10.3390/su6031313
APA StyleMcLain, J. E., & Williams, C. F. (2014). Sustainability of Water Reclamation: Long-Term Recharge with Reclaimed Wastewater Does Not Enhance Antibiotic Resistance in Sediment Bacteria. Sustainability, 6(3), 1313-1327. https://doi.org/10.3390/su6031313