Estimating Forest Leaf Area Index and Canopy Chlorophyll Content with Sentinel-2: An Evaluation of Two Hybrid Retrieval Algorithms
Abstract
:1. Introduction
- Determine the retrieval accuracy that can be expected when the SNAP L2B retrieval algorithm (which is based on SAIL and not optimised for forest environments) is used for LAI and CCC retrieval over deciduous broadleaf forest.
- Evaluate the extent to which a retrieval algorithm trained using INFORM (and thus optimised for forest environments) will improve LAI and CCC retrieval accuracy.
2. Materials and Methods
2.1. Study Site
2.2. Field Data Collection and Processing
2.3. Interpolation of Field Data
2.4. MSI Data Pre-Processing
2.5. Hybrid LAI and CCC Retrieval
2.6. Forward Modelling Experiments
2.7. Performance Metrics
3. Results
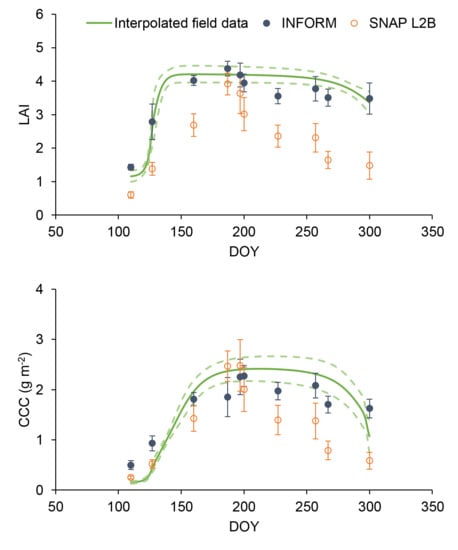
3.1. Field Data and Interpolation
3.2. Overall Performance of the Retrieval Algorithms
3.3. Phenological Variations in Performance
3.4. Reproduction of Observed MSI Spectra by SAIL and INFORM
4. Discussion
4.1. Utility of Interpolating Field Data
4.2. Choice of Retrieval Algorithms for Forest-Related Applications
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| ANN | LAI | CCC | ||||
|---|---|---|---|---|---|---|
| r2 | RMSE | NRMSE (%) | r2 | RMSE | NRMSE (%) | |
| 1 | 0.58 | 0.45 | 11.36 | 0.60 | 0.40 | 20.25 |
| 2 | 0.59 | 0.45 | 11.41 | 0.61 | 0.40 | 20.09 |
| 3 | 0.58 | 0.45 | 11.43 | 0.61 | 0.40 | 20.17 |
| 4 | 0.58 | 0.45 | 11.35 | 0.61 | 0.40 | 20.11 |
| 5 | 0.59 | 0.45 | 11.32 | 0.61 | 0.40 | 20.20 |
| 6 | 0.58 | 0.45 | 11.47 | 0.59 | 0.41 | 20.60 |
| 7 | 0.59 | 0.44* | 11.27 | 0.61 | 0.40 | 20.22 |
| 8 | 0.58 | 0.45 | 11.42 | 0.62 | 0.40 | 20.13 |
| 9 | 0.59 | 0.45 | 11.37 | 0.60 | 0.40 | 20.35 |
| 10 | 0.58 | 0.45 | 11.45 | 0.61 | 0.39* | 20.00 |
References
- Running, S.W.; Nemani, R.R.; Heinsch, F.A.; Zhao, M.; Reeves, M.; Hashimoto, H. A Continuous Satellite-Derived Measure of Global Terrestrial Primary Production. Bioscience 2004, 54, 547. [Google Scholar] [CrossRef]
- Sellers, P.J.; Tucker, C.J.; Collatz, G.J.; Los, S.O.; Justice, C.O.; Dazlich, D.A.; Randall, D.A. A Revised Land Surface Parameterization (SiB2) for Atmospheric GCMS. Part II: The Generation of Global Fields of Terrestrial Biophysical Parameters from Satellite Data. J. Clim. 1996, 9, 706–737. [Google Scholar] [CrossRef] [Green Version]
- Sellers, P.J.; Dickinson, R.E.; Randall, D.A.; Betts, A.K.; Hall, F.G.; Berry, J.A.; Collatz, G.J.; Denning, A.S.; Mooney, H.A.; Nobre, C.A.; et al. Modeling the Exchanges of Energy, Water, and Carbon Between Continents and the Atmosphere. Science 1997, 275, 502–509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beer, C.; Reichstein, M.; Tomelleri, E.; Ciais, P.; Jung, M.; Carvalhais, N.; Rodenbeck, C.; Arain, M.A.; Baldocchi, D.; Bonan, G.B.; et al. Terrestrial Gross Carbon Dioxide Uptake: Global Distribution and Covariation with Climate. Science. 2010, 329, 834–838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- FAO Global Forest Resources Assessment 2015; Food and Agriculture Organizaion of the United Nations: Rome, Italy, 2015; ISBN 978-92-5-108826-5.
- Barredo, J.I.; Bastrup-Birk, A.; Teller, A.; Onaindia, M.; Fernández de Manuel, B.; Madariaga, I.; Rodríguez-Loinaz, G.; Pinho, P.; Nunes, A.; Ramos, A.; et al. Mapping and Assessment of Forest Ecosystems and Their Services—Applications and Guidance for Decision Making in the Framework of MAES; European Commission Joint Research Centre: Ispra, Italy, 2015; ISBN 978-92-79-55332-5. [Google Scholar]
- Bréda, N.J.J. Ground-based measurements of leaf area index: A review of methods, instruments and current controversies. J. Exp. Bot. 2003, 54, 2403–2417. [Google Scholar] [CrossRef]
- Jonckheere, I.; Fleck, S.; Nackaerts, K.; Muys, B.; Coppin, P.; Weiss, M.; Baret, F. Review of methods for in situ leaf area index determination. Agric. For. Meteorol. 2004, 121, 19–35. [Google Scholar] [CrossRef]
- Markwell, J.; Osterman, J.C.; Mitchell, J.L. Calibration of the Minolta SPAD-502 leaf chlorophyll meter. Photosynth. Res. 1995, 46, 467–472. [Google Scholar] [CrossRef]
- Verrelst, J.; Camps-Valls, G.; Muñoz-Marí, J.; Rivera, J.P.; Veroustraete, F.; Clevers, J.G.P.W.; Moreno, J. Optical remote sensing and the retrieval of terrestrial vegetation bio-geophysical properties—A review. ISPRS J. Photogramm. Remote Sens. 2015, 108, 273–290. [Google Scholar] [CrossRef]
- Liang, S. Recent developments in estimating land surface biogeophysical variables from optical remote sensing. Prog. Phys. Geogr. 2007, 31, 501–516. [Google Scholar] [CrossRef] [Green Version]
- Richter, K.; Hank, T.B.; Vuolo, F.; Mauser, W.; D’Urso, G. Optimal Exploitation of the Sentinel-2 Spectral Capabilities for Crop Leaf Area Index Mapping. Remote Sens. 2012, 4, 561–582. [Google Scholar] [CrossRef] [Green Version]
- Knyazikhin, Y.; Martonchik, J.V.; Myneni, R.B.; Diner, D.J.; Running, S.W. Synergistic algorithm for estimating vegetation canopy leaf area index and fraction of absorbed photosynthetically active radiation from MODIS and MISR data. J. Geophys. Res. Atmos. 1998, 103, 32257–32275. [Google Scholar] [CrossRef] [Green Version]
- Myneni, R.B.; Hoffman, S.; Knyazikhin, Y.; Privette, J.L.; Glassy, J.; Tian, Y.; Wang, Y.; Song, X.; Zhang, Y.; Smith, G.R.; et al. Global products of vegetation leaf area and fraction absorbed PAR from year one of MODIS data. Remote Sens. Environ. 2002, 83, 214–231. [Google Scholar] [CrossRef] [Green Version]
- Baret, F.; Weiss, M.; Lacaze, R.; Camacho, F.; Makhmara, H.; Pacholcyzk, P.; Smets, B. GEOV1: LAI and FAPAR essential climate variables and FCOVER global time series capitalizing over existing products. Part 1: Principles of development and production. Remote Sens. Environ. 2013, 137, 299–309. [Google Scholar] [CrossRef]
- Baret, F.; Hagolle, O.; Geiger, B.; Bicheron, P.; Miras, B.; Huc, M.; Berthelot, B.; Niño, F.; Weiss, M.; Samain, O.; et al. LAI, fAPAR and fCover CYCLOPES global products derived from VEGETATION. Remote Sens. Environ. 2007, 110, 275–286. [Google Scholar] [CrossRef]
- Lacaze, R.; Smets, B.; Baret, F.; Weiss, M.; Ramon, D.; Montersleet, B.; Wandrebeck, L.; Calvet, J.-C.; Roujean, J.-L.; Camacho, F. OPERATIONAL 333m BIOPHYSICAL PRODUCTS OF THE COPERNICUS GLOBAL LAND SERVICE FOR AGRICULTURE MONITORING. ISPRS - Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2015, XL-7/W3, 53–56. [Google Scholar] [CrossRef] [Green Version]
- Vuolo, F.; Atzberger, C.; Richter, K.; Dash, J. Retrieval of Biophysical Vegetation Products From Rapideye Imagery. In Proceedings of the ISPRS TC VII Symposium, Vienna, Austria, 5–7 July 2010; Wagner, W., Székely, B., Eds.; International Society for Photogrammetry and Remote Sensing: Hannover, Germany; pp. 281–286.
- Verrelst, J.; Rivera, J.P.; Veroustraete, F.; Muñoz-Marí, J.; Clevers, J.G.P.W.; Camps-Valls, G.; Moreno, J. Experimental Sentinel-2 LAI estimation using parametric, non-parametric and physical retrieval methods – A comparison. ISPRS J. Photogramm. Remote Sens. 2015, 108, 260–272. [Google Scholar] [CrossRef]
- Verrelst, J.; Muñoz, J.; Alonso, L.; Delegido, J.; Rivera, J.P.; Camps-Valls, G.; Moreno, J. Machine learning regression algorithms for biophysical parameter retrieval: Opportunities for Sentinel-2 and -3. Remote Sens. Environ. 2012, 118, 127–139. [Google Scholar] [CrossRef]
- Bacour, C.; Baret, F.; Béal, D.; Weiss, M.; Pavageau, K. Neural network estimation of LAI, fAPAR, fCover and LAIxCab, from top of canopy MERIS reflectance data: Principles and validation. Remote Sens. Environ. 2006, 105, 313–325. [Google Scholar] [CrossRef]
- Kimes, D.S.; Knyazikhin, Y.; Privette, J.L.; Abuelgasim, A.A.; Gao, F. Inversion methods for physically-based models. Remote Sens. Rev. 2000, 18, 381–439. [Google Scholar] [CrossRef]
- Verger, A.; Baret, F.; Camacho, F. Optimal modalities for radiative transfer-neural network estimation of canopy biophysical characteristics: Evaluation over an agricultural area with CHRIS/PROBA observations. Remote Sens. Environ. 2011, 115, 415–426. [Google Scholar] [CrossRef]
- Drusch, M.; Del Bello, U.; Carlier, S.; Colin, O.; Fernandez, V.; Gascon, F.; Hoersch, B.; Isola, C.; Laberinti, P.; Martimort, P.; et al. Sentinel-2: ESA’s Optical High-Resolution Mission for GMES Operational Services. Remote Sens. Environ. 2012, 120, 25–36. [Google Scholar] [CrossRef]
- Clevers, J.G.P.W.; Gitelson, A.A. Remote estimation of crop and grass chlorophyll and nitrogen content using red-edge bands on Sentinel-2 and -3. Int. J. Appl. Earth Obs. Geoinf. 2013, 23, 344–351. [Google Scholar] [CrossRef]
- Clevers, J.; Kooistra, L.; van den Brande, M. Using Sentinel-2 Data for Retrieving LAI and Leaf and Canopy Chlorophyll Content of a Potato Crop. Remote Sens. 2017, 9, 405. [Google Scholar] [CrossRef]
- Frampton, W.J.; Dash, J.; Watmough, G.; Milton, E.J. Evaluating the capabilities of Sentinel-2 for quantitative estimation of biophysical variables in vegetation. ISPRS J. Photogramm. Remote Sens. 2013, 82, 83–92. [Google Scholar] [CrossRef] [Green Version]
- Darvishzadeh, R.; Skidmore, A.; Abdullah, H.; Cherenet, E.; Ali, A.; Wang, T.; Nieuwenhuis, W.; Heurich, M.; Vrieling, A.; O’Connor, B.; et al. Mapping leaf chlorophyll content from Sentinel-2 and RapidEye data in spruce stands using the invertible forest reflectance model. Int. J. Appl. Earth Obs. Geoinf. 2019, 79, 58–70. [Google Scholar] [CrossRef] [Green Version]
- Inoue, Y.; Guérif, M.; Baret, F.; Skidmore, A.; Gitelson, A.; Schlerf, M.; Darvishzadeh, R.; Olioso, A. Simple and robust methods for remote sensing of canopy chlorophyll content: A comparative analysis of hyperspectral data for different types of vegetation. Plant. Cell Environ. 2016, 39, 2609–2623. [Google Scholar] [CrossRef] [PubMed]
- Gholizadeh, A.; Mišurec, J.; Kopačková, V.; Mielke, C.; Rogass, C. Assessment of Red-Edge Position Extraction Techniques: A Case Study for Norway Spruce Forests Using HyMap and Simulated Sentinel-2 Data. Forests 2016, 7, 226. [Google Scholar] [CrossRef]
- Majasalmi, T.; Rautiainen, M. The potential of Sentinel-2 data for estimating biophysical variables in a boreal forest: A simulation study. Remote Sens. Lett. 2016, 7, 427–436. [Google Scholar] [CrossRef]
- Korhonen, L.; Hadi; Packalen, P.; Rautiainen, M. Comparison of Sentinel-2 and Landsat 8 in the estimation of boreal forest canopy cover and leaf area index. Remote Sens. Environ. 2017, 195, 259–274. [Google Scholar] [CrossRef]
- Weiss, M.; Baret, F. S2ToolBox Level 2 Products: LAI, FAPAR, FCOVER; 1.1.; Institut National de la Recherche Agronomique: Avignon, France, 2016. [Google Scholar]
- Verhoef, W.; Jia, L.; Xiao, Q.; Su, Z. Unified Optical-Thermal Four-Stream Radiative Transfer Theory for Homogeneous Vegetation Canopies. IEEE Trans. Geosci. Remote Sens. 2007, 45, 1808–1822. [Google Scholar] [CrossRef]
- Feret, J.-B.; François, C.; Asner, G.P.; Gitelson, A.A.; Martin, R.E.; Bidel, L.P.R.; Ustin, S.L.; le Maire, G.; Jacquemoud, S. PROSPECT-4 and 5: Advances in the leaf optical properties model separating photosynthetic pigments. Remote Sens. Environ. 2008, 112, 3030–3043. [Google Scholar] [CrossRef]
- Vuolo, F.; Żółtak, M.; Pipitone, C.; Zappa, L.; Wenng, H.; Immitzer, M.; Weiss, M.; Baret, F.; Atzberger, C. Data Service Platform for Sentinel-2 Surface Reflectance and Value-Added Products: System Use and Examples. Remote Sens. 2016, 8, 938. [Google Scholar] [CrossRef]
- Djamai, N.; Fernandes, R.; Weiss, M.; McNairn, H.; Goïta, K. Validation of the Sentinel Simplified Level 2 Product Prototype Processor (SL2P) for mapping cropland biophysical variables using Sentinel-2/MSI and Landsat-8/OLI data. Remote Sens. Environ. 2019, 225, 416–430. [Google Scholar] [CrossRef]
- Xie, Q.; Dash, J.; Huete, A.; Jiang, A.; Yin, G.; Ding, Y.; Peng, D.; Hall, C.C.; Brown, L.; Shi, Y.; et al. Retrieval of crop biophysical parameters from Sentinel-2 remote sensing imagery. Int. J. Appl. Earth Obs. Geoinf. 2019, 80, 187–195. [Google Scholar] [CrossRef]
- Vanino, S.; Nino, P.; De Michele, C.; Falanga Bolognesi, S.; D’Urso, G.; Di Bene, C.; Pennelli, B.; Vuolo, F.; Farina, R.; Pulighe, G.; et al. Capability of Sentinel-2 data for estimating maximum evapotranspiration and irrigation requirements for tomato crop in Central Italy. Remote Sens. Environ. 2018, 215, 452–470. [Google Scholar] [CrossRef]
- Schlerf, M.; Atzberger, C. Inversion of a forest reflectance model to estimate structural canopy variables from hyperspectral remote sensing data. Remote Sens. Environ. 2006, 100, 281–294. [Google Scholar] [CrossRef]
- Dash, J.; Almond, S.F.; Boyd, D.; Curran, P.J. Multi-scale analysis and validation of the Envisat MERIS Terrestrial Chlorophyll Index (MTCI) in woodland. In Proceedings of the 2nd MERIS/(A)ATSR User Workshop, Frascati, Italy, 22–26 September 2008; European Space Agency: Noordwijk, The Netherlands.
- Ogutu, B.; Dash, J.; Dawson, T.P. Evaluation of leaf area index estimated from medium spatial resolution remote sensing data in a broadleaf deciduous forest in southern England, UK. Can. J. Remote Sens. 2012, 37, 333–347. [Google Scholar] [CrossRef]
- Cantarello, E.; Newton, A.C. Identifying cost-effective indicators to assess the conservation status of forested habitats in Natura 2000 sites. For. Ecol. Manage. 2008, 256, 815–826. [Google Scholar] [CrossRef]
- Mountford, E.P.; Peterken, G.F.; Edwards, P.J.; Manners, J.G. Long-term change in growth, mortality and regeneration of trees in Denny Wood, an old-growth wood-pasture in the New Forest (UK). Perspect. Plant Ecol. Evol. Syst. 1999, 2, 223–272. [Google Scholar] [CrossRef]
- Justice, C.O.; Townshend, J.R.G. Integrating ground data with remote sensing. In Terrain Analysis and Remote Sensing; Townshend, J.R.G., Ed.; Allen and Unwin: London, UK, 1981; pp. 35–58. [Google Scholar]
- Gascon, F.; Bouzinac, C.; Thépaut, O.; Jung, M.; Francesconi, B.; Louis, J.; Lonjou, V.; Lafrance, B.; Massera, S.; Gaudel-Vacaresse, A.; et al. Copernicus Sentinel-2A Calibration and Products Validation Status. Remote Sens. 2017, 9, 584. [Google Scholar] [CrossRef]
- Garmin. Garmin eTrex H Owner’s Manual; Garmin: Olathe, KS, USA, 2007; ISBN 0808238000. [Google Scholar]
- Campbell, J.L.; Burrows, S.; Gower, S.T.; Cohen, W.B. BigFoot: Characterizing Land Cover, LAI and NPP at the Landscape Scale for EOS/MODIS Validation - Field Manual; 2.1.; Oak Ridge National Laboratory: Oak Ridge, TN, USA, 1999; ISBN 0071601201. [Google Scholar]
- Weiss, M.; Baret, F. CAN-EYE V6.4.91 User Manual; Institut National de la Recherche Agronomique: Avignon, France, 2017. [Google Scholar]
- Miller, J. A formula for average foliage density. Aust. J. Bot. 1967, 15, 141–144. [Google Scholar] [CrossRef]
- Lang, A.R.G.; Yueqin, X. Estimation of leaf area index from transmission of direct sunlight in discontinuous canopies. Agric. For. Meteorol. 1986, 37, 229–243. [Google Scholar] [CrossRef]
- Weiss, M.; Baret, F.; Smith, G.J.; Jonckheere, I.; Coppin, P. Review of methods for in situ leaf area index (LAI) determination Part II: Estimation of LAI, errors and sampling. Agric. For. Meteorol. 2004, 121, 37–53. [Google Scholar] [CrossRef]
- Demarez, V. Seasonal variation of leaf chlorophyll content of a temperate forest. Inversion of the PROSPECT model. Int. J. Remote Sens. 1999, 20, 879–894. [Google Scholar] [CrossRef]
- Uddling, J.; Gelang-Alfredsson, J.; Piikki, K.; Pleijel, H. Evaluating the relationship between leaf chlorophyll concentration and SPAD-502 chlorophyll meter readings. Photosynth. Res. 2007, 91, 37–46. [Google Scholar] [CrossRef]
- Brown, L.A.; Dash, J.; Lidón, A.L.; Lopez-Baeza, E.; Dransfeld, S. Synergetic Exploitation of the Sentinel-2 Missions for Validating the Sentinel-3 Ocean and Land Colour Instrument Terrestrial Chlorophyll Index over a Vineyard Dominated Mediterranean Environment. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2019, 12. [Google Scholar] [CrossRef]
- Fernandes, R.; Plummer, S.; Nightingale, J.; Baret, F.; Camacho, F.; Fang, H.; Garrigues, S.; Gobron, N.; Lang, M.; Lacaze, R.; et al. Global Leaf Area Index Product Validation Good Practices. In Best Practice for Satellite-Derived Land Product Validation; Fernandes, R., Plummer, S., Nightingale, J., Eds.; Land Product Validation Subgroup (Committee on Earth Observation Satellites Working Group on Calibration and Validation): Greenbelt, MD, USA, 2014. [Google Scholar]
- Morisette, J.T.; Baret, F.; Privette, J.L.; Myneni, R.B.; Nickeson, J.E.; Garrigues, S.; Shabanov, N.V.; Weiss, M.; Fernandes, R.A.; Leblanc, S.G.; et al. Validation of global moderate-resolution LAI products: A framework proposed within the CEOS land product validation subgroup. IEEE Trans. Geosci. Remote Sens. 2006, 44, 1804–1817. [Google Scholar] [CrossRef]
- Vuolo, F.; Dash, J.; Curran, P.J.; Lajas, D.; Kwiatkowska, E. Methodologies and Uncertainties in the Use of the Terrestrial Chlorophyll Index for the Sentinel-3 Mission. Remote Sens. 2012, 4, 1112–1133. [Google Scholar] [CrossRef] [Green Version]
- Baret, F.; Weiss, M.; Allard, D.; Garrigues, S.; Leroy, M.; Jeanjean, H.; Fernandes, R.; Myneni, R.; Privette, J.; Morisette, J.; et al. VALERI: A Network of Sites and a Methodology for the Validation of Medium Spatial Resolution Land Satellite Products; Institut National de la Recherche Agronomique: Avignon, France, 2005. [Google Scholar]
- De Kauwe, M.G.; Disney, M.I.; Quaife, T.; Lewis, P.; Williams, M. An assessment of the MODIS collection 5 leaf area index product for a region of mixed coniferous forest. Remote Sens. Environ. 2011, 115, 767–780. [Google Scholar] [CrossRef]
- Atkinson, P.M.; Jeganathan, C.; Dash, J.; Atzberger, C. Inter-comparison of four models for smoothing satellite sensor time-series data to estimate vegetation phenology. Remote Sens. Environ. 2012, 123, 400–417. [Google Scholar] [CrossRef]
- Beck, P.S.A.; Atzberger, C.; Høgda, K.A.; Johansen, B.; Skidmore, A.K. Improved monitoring of vegetation dynamics at very high latitudes: A new method using MODIS NDVI. Remote Sens. Environ. 2006, 100, 321–334. [Google Scholar] [CrossRef]
- Zhang, X.; Friedl, M.A.; Schaaf, C.B.; Strahler, A.H.; Hodges, J.C.F.; Gao, F.; Reed, B.C.; Huete, A. Monitoring vegetation phenology using MODIS. Remote Sens. Environ. 2003, 84, 471–475. [Google Scholar] [CrossRef]
- Müller-Wilm, U. Sentinel-2 MSI – Level-2A Prototype Processor Installation and User Manual; Telespazio VEGA: Darmstadt, Germany, 2016. [Google Scholar]
- ESA SNAP. Available online: http://step.esa.int/main/toolboxes/snap/ (accessed on 24 August 2018).
- Weiss, M.; Baret, F.; Myneni, R.B.; Pragnère, A.; Knyazikhin, Y. Investigation of a model inversion technique to estimate canopy biophysical variables from spectral and directional reflectance data. Agronomie 2000, 20, 3–22. [Google Scholar] [CrossRef]
- Combal, B.; Baret, F.; Weiss, M.; Trubuil, A.; Macé, D.; Pragnère, A.; Myneni, R.; Knyazikhin, Y.; Wang, L. Retrieval of canopy biophysical variables from bidirectional reflectance. Remote Sens. Environ. 2003, 84, 1–15. [Google Scholar] [CrossRef]
- Baldridge, A.M.; Hook, S.J.; Grove, C.I.; Rivera, G. The ASTER spectral library version 2.0. Remote Sens. Environ. 2009, 113, 711–715. [Google Scholar] [CrossRef]
- ESA Sentinel-2 Spectral Response Functions (S2-SRF). Available online: https://earth.esa.int/documents/247904/685211/Sentinel-2+MSI+Spectral+Responses/ (accessed on 16 May 2017).
- Li, W.; Weiss, M.; Waldner, F.; Defourny, P.; Demarez, V.; Morin, D.; Hagolle, O.; Baret, F. A Generic Algorithm to Estimate LAI, FAPAR and FCOVER Variables from SPOT4_HRVIR and Landsat Sensors: Evaluation of the Consistency and Comparison with Ground Measurements. Remote Sens. 2015, 7, 15494–15516. [Google Scholar] [CrossRef] [Green Version]
- Upreti, D.; Huang, W.; Kong, W.; Pascucci, S.; Pignatti, S.; Zhou, X.; Ye, H.; Casa, R. A Comparison of Hybrid Machine Learning Algorithms for the Retrieval of Wheat Biophysical Variables from Sentinel-2. Remote Sens. 2019, 11, 481. [Google Scholar] [CrossRef]
- Schlerf, M.; Atzberger, C. Vegetation Structure Retrieval in Beech and Spruce Forests Using Spectrodirectional Satellite Data. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2012, 5, 8–17. [Google Scholar] [CrossRef]
- Yuan, H.; Ma, R.; Atzberger, C.; Li, F.; Loiselle, S.; Luo, J. Estimating Forest fAPAR from Multispectral Landsat-8 Data Using the Invertible Forest Reflectance Model INFORM. Remote Sens. 2015, 7, 7425–7446. [Google Scholar] [CrossRef] [Green Version]
- Atzberger, C.; Darvishzadeh, R.; Schlerf, M.; Le Maire, G. Suitability and adaptation of PROSAIL radiative transfer model for hyperspectral grassland studies. Remote Sens. Lett. 2013, 4, 55–64. [Google Scholar] [CrossRef]
- Darvishzadeh, R.; Atzberger, C.; Skidmore, A.; Schlerf, M. Mapping grassland leaf area index with airborne hyperspectral imagery: A comparison study of statistical approaches and inversion of radiative transfer models. ISPRS J. Photogramm. Remote Sens. 2011, 66, 894–906. [Google Scholar] [CrossRef]
- Darvishzadeh, R.; Skidmore, A.; Schlerf, M.; Atzberger, C. Inversion of a radiative transfer model for estimating vegetation LAI and chlorophyll in a heterogeneous grassland. Remote Sens. Environ. 2008, 112, 2592–2604. [Google Scholar] [CrossRef]
- Richter, K.; Atzberger, C.; Vuolo, F.; Weihs, P.; D’Urso, G. Experimental assessment of the Sentinel-2 band setting for RTM-based LAI retrieval of sugar beet and maize. Can. J. Remote Sens. 2009, 35, 230–247. [Google Scholar] [CrossRef]
- Verrelst, J.; Rivera, J.P.; Gitelson, A.; Delegido, J.; Moreno, J.; Camps-Valls, G. Spectral band selection for vegetation properties retrieval using Gaussian processes regression. Int. J. Appl. Earth Obs. Geoinf. 2016, 52, 554–567. [Google Scholar] [CrossRef]
- Heiskanen, J.; Rautiainen, M.; Korhonen, L.; Mõttus, M.; Stenberg, P. Retrieval of boreal forest LAI using a forest reflectance model and empirical regressions. Int. J. Appl. Earth Obs. Geoinf. 2011, 13, 595–606. [Google Scholar] [CrossRef]
- Heiskanen, J.; Rautiainen, M.; Stenberg, P.; Mõttus, M.; Vesanto, V.-H.; Korhonen, L.; Majasalmi, T. Seasonal variation in MODIS LAI for a boreal forest area in Finland. Remote Sens. Environ. 2012, 126, 104–115. [Google Scholar] [CrossRef]
- Baltzer, J.L.; Thomas, S.C. Leaf optical responses to light and soil nutrient availability in temperate deciduous trees. Am. J. Bot. 2005, 92, 214–223. [Google Scholar] [CrossRef]





| Stand Attribute | Minimum | Maximum | Mean | Standard Deviation |
|---|---|---|---|---|
| Canopy height (m) | 10 | 34 | - | - |
| Diameter at breast height (cm) | 25.9 | 80.5 | 43.4 | 12.9 |
| Crown diameter (m) | 5.76 | 14.29 | 8.49 | 3.73 |
| Stem density (ha−1) | 72 | 536 | 256 | 106 |
| Parameter | Minimum | Maximum | Mean | Standard Deviation | Distribution | Reference |
|---|---|---|---|---|---|---|
| Structural parameter (N) | 1.5 | 1.7 | - | - | Uniform | [72,73] |
| Chlorophyll a + b (µg cm−2) | 10 | 60 | 50 | 20 | Gaussian | This study |
| Dry matter (g cm−2) | 0.004 | 0.02 | - | - | Uniform | [72,73] |
| Equivalent water thickness (g cm−2) | 0.01 | 0.02 | - | - | Uniform | [72,73] |
| Average leaf angle (°) | 55 | 55 | - | - | Fixed | [72,73] |
| Single tree LAI | 1.0 | 5.0 | 4.0 | 0.5 | Gaussian | This study |
| Understory LAI | 0.5 | 0.5 | - | - | Fixed | [72,73] |
| Stem density (ha−1) | 72 | 536 | 256 | 106 | Gaussian | [43] |
| Canopy height (m) | 10 | 34 | - | - | Uniform | [44] |
| Crown diameter (m) | 6 | 14 | 8 | 4 | Gaussian | [43] |
| Solar zenith angle (°) | 29 | 64 | - | - | Uniform | This study |
| Observer zenith angle (°) | 3 | 11 | - | - | Uniform | This study |
| Relative azimuth angle (°) | 20 | 136 | - | - | Uniform | This study |
| Soil brightness coefficient | 0.5 | 0.5 | - | - | Fixed | [72] |
| Fraction of diffuse radiation | 0.1 | 0.1 | - | - | Fixed | [40,72,73] |
| Band | Central Wavelength (nm) | Bandwidth (nm) | Native Spatial Resolution (m) |
|---|---|---|---|
| B3 | 550 | 35 | 10 |
| B4 | 665 | 30 | 10 |
| B5 | 705 | 15 | 20 |
| B6 | 740 | 15 | 20 |
| B7 | 783 | 20 | 20 |
| B8A | 865 | 20 | 20 |
| B11 | 1610 | 90 | 20 |
| B12 | 2190 | 180 | 20 |
| LAI | LCC | ||
|---|---|---|---|
| DOY | Absolute Error | DOY | Absolute Error (g m−2) |
| 101 | 0.02 | 120 | 0.21 |
| 112 | 0.00 | 147 | 0.01 |
| 126 | 0.10 | 160 | 0.03 |
| 140 | 0.16 | 176 | 0.03 |
| 155 | 0.04 | 208 | 0.04 |
| 197 | 0.05 | 259 | 0.04 |
| 278 | 0.04 | 281 | 0.05 |
| 307 | 0.18 | 300 | 0.22 |
| Variable | DOY | SNAP L2B | INFORM | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| r2 | RMSE (NRMSE) | Bias | Precision | n | r2 | RMSE (NRMSE) | Bias | Precision | n | ||
| LAI | 100 to 149 | 0.77 | 1.22 (60.28%) | −1.03 | 0.68 | 17 | 0.74 | 0.51 (25.05%) | 0.09 | 0.51 | 18 |
| 150 to 249 | 0.07 | 1.34 (32.06%) | −1.17 | 0.66 | 39 | 0.11 | 0.40 (9.61%) | −0.19 | 0.36 | 43 | |
| 250 to 300 | 0.14 | 2.00 (52.76%) | −1.94 | 0.49 | 25 | 0.02 | 0.55 (14.47%) | −0.18 | 0.53 | 26 | |
| CCC | 100 to 149 | 0.70 | 0.12 (36.78%) | 0.06 | 0.11 | 17 | 0.66 | 0.41 (126.32%) | 0.39 | 0.15 | 18 |
| 150 to 249 | 0.10 | 0.72 (30.89%) | −0.45 | 0.57 | 39 | 0.04 | 0.49 (20.94%) | −0.30 | 0.39 | 43 | |
| 250 to 300 | 0.26 | 1.11 (59.92%) | −0.94 | 0.59 | 25 | 0.17 | 0.62 (33.56%) | −0.04 | 0.63 | 26 | |
| Band | SAIL | INFORM | ||||||
|---|---|---|---|---|---|---|---|---|
| r2 | RMSE (NRMSE) | Bias | Precision | r2 | RMSE (NRMSE) | Bias | Precision | |
| B3 | 0.06 | 0.03 (56.15%) | −0.01 | 0.03 | 0.23 | 0.02 (39.68%) | −0.01 | 0.02 |
| B4 | 0.15 | 0.03 (72.92%) | −0.02 | 0.02 | 0.12 | 0.02 (58.91%) | −0.01 | 0.02 |
| B5 | 0.21 | 0.04 (38.68%) | −0.02 | 0.03 | 0.43 | 0.03 (32.60%) | −0.02 | 0.02 |
| B6 | 0.84 | 0.04 (16.95%) | 0.02 | 0.04 | 0.96 | 0.02 (6.86%) | 0.00 | 0.02 |
| B7 | 0.92 | 0.04 (13.60%) | 0.03 | 0.03 | 0.99 | 0.01 (4.72%) | 0.01 | 0.01 |
| B8A | 0.93 | 0.04 (10.31%) | 0.00 | 0.04 | 0.99 | 0.01 (3.68%) | −0.01 | 0.01 |
| B11 | 0.73 | 0.02 (11.36%) | −0.01 | 0.02 | 0.78 | 0.02 (11.68%) | −0.01 | 0.02 |
| B12 | 0.55 | 0.03 (37.65%) | −0.03 | 0.02 | 0.63 | 0.02 (25.88%) | −0.01 | 0.02 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brown, L.A.; Ogutu, B.O.; Dash, J. Estimating Forest Leaf Area Index and Canopy Chlorophyll Content with Sentinel-2: An Evaluation of Two Hybrid Retrieval Algorithms. Remote Sens. 2019, 11, 1752. https://doi.org/10.3390/rs11151752
Brown LA, Ogutu BO, Dash J. Estimating Forest Leaf Area Index and Canopy Chlorophyll Content with Sentinel-2: An Evaluation of Two Hybrid Retrieval Algorithms. Remote Sensing. 2019; 11(15):1752. https://doi.org/10.3390/rs11151752
Chicago/Turabian StyleBrown, Luke A., Booker O. Ogutu, and Jadunandan Dash. 2019. "Estimating Forest Leaf Area Index and Canopy Chlorophyll Content with Sentinel-2: An Evaluation of Two Hybrid Retrieval Algorithms" Remote Sensing 11, no. 15: 1752. https://doi.org/10.3390/rs11151752
APA StyleBrown, L. A., Ogutu, B. O., & Dash, J. (2019). Estimating Forest Leaf Area Index and Canopy Chlorophyll Content with Sentinel-2: An Evaluation of Two Hybrid Retrieval Algorithms. Remote Sensing, 11(15), 1752. https://doi.org/10.3390/rs11151752