Looking for Ticks from Space: Using Remotely Sensed Spectral Diversity to Assess Amblyomma and Hyalomma Tick Abundance
Abstract
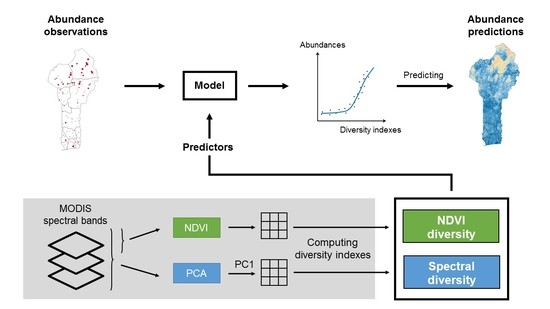
:1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Sampling Protocol
2.3. Tick Autecology
2.4. Satellite Images and Predictor Variables
2.5. Data Analysis
3. Results
3.1. Land Cover Analysis
3.2. Poisson Zero-Inflated Models
4. Discussion
4.1. Ecological Perspectives of Spectral Diversity Measures
4.2. Limits of the Study and Future Perspective
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zeman, P.; Lynen, G. Conditions for stable parapatric coexistence between Boophilusdecoloratus and B. microplus ticks: A simulation study using the competitive Lotka-Volterra model. Exp. Appl. Acarol. 2010, 52, 409–426. [Google Scholar] [CrossRef] [PubMed]
- Escobar, L.E.; Craft, M.E. Advances and limitations of disease biogeography using ecological niche modeling. Front. Microbiol. 2016, 7, 1174. [Google Scholar] [CrossRef] [PubMed]
- Medlock, J.M.; Hansford, K.M.; Bormane, A.; Derdakova, M.; Estrada-Peña, A.; George, J.C.; Golovljova, I.; Jaenson, T.G.T.; Jensen, J.K.; Jensen, P.M.; et al. Driving forces for changes in geographical distribution of Ixodes ricinus ticks in Europe. Parasit. Vectors 2013, 6, 1. [Google Scholar] [CrossRef]
- Dautel, H.; Knülle, W. Cold hardiness, supercooling ability and causes of low-temperature mortality in the soft tick, Argas reflexus, and the hard tick, Ixodes ricinus (Acari: Ixodoidea) from Central Europe. J. Insect Physiol. 1997, 43, 843–854. [Google Scholar] [CrossRef]
- Pfäffle, M.; Littwin, N.; Muders, S.V.; Petney, T.N. The ecology of tick-borne diseases. Int. J. Parasitol. 2013, 43, 1059–1077. [Google Scholar] [CrossRef]
- Bowman, A.S.; Nuttall, P.A. Ticks: Biology, Disease and Control; Cambridge University Press: Cambridge, UK, 2008. [Google Scholar]
- Kalluri, S.; Gilruth, P.; Rogers, D.; Szczur, M. Surveillance of arthropod vector-borne infectious diseases using remote sensing techniques: A review. PLoS Pathog. 2007, 3, e116. [Google Scholar] [CrossRef]
- Rocchini, D.; Boyd, D.S.; Féret, J.B.; Foody, G.M.; He, K.S.; Lausch, A.; Nagendra, H.; Wegmann, M.; Pettorelli, N. Satellite remote sensing to monitor species diversity: Potential and pitfalls. Remote Sens. Ecolo. Conserv. 2016, 2, 25–36. [Google Scholar] [CrossRef]
- Rogers, D.J.; Hay, S.I.; & Packer, M.J. Predicting the distribution of tsetse flies in West Africa using temporal Fourier processed meteorological satellite data. Ann. Trop. Med. Parasitol. 1996, 90, 225–241. [Google Scholar] [CrossRef]
- Randolph, S.E. Abiotic and biotic determinants of the seasonal dynamics of the tick Rhipicephalus appendiculatus in South Africa. Med. Vet. Entomol. 1997, 11, 25–37. [Google Scholar] [CrossRef]
- Ogden, N.H.; Trudel, L.; Artsob, H.; Barker, I.K.; Beauchamp, G.; Charron, D.; Drebot, M.A.; Galloway, T.D.; O’Handley, R.; Thompson, R.A.; et al. Ixodes scapularis ticks collected by passive surveillance in Canada: Analysis of geographic distribution and infection with the Lyme borreliosis agent Borrelia burgdorferi. J. Med. Entomol. 2006, 43, 600–609. [Google Scholar] [CrossRef]
- Altobelli, A.; Boemo, B.; Mignozzi, K.; Bandi, M.; Floris, R.; Menardi, G.; Cinco, M. Spatial Lyme borreliosis risk assessment in north-eastern Italy. Int. J. Med. Microbiol. 2008, 298, 125–128. [Google Scholar] [CrossRef]
- Palmer, M.W.; Earls, P.G.; Hoagland, B.W.; White, P.S.; Wohlgemuth, T. Quantitative tools for perfecting species lists. Environmetrics 2002, 13, 121–137. [Google Scholar] [CrossRef]
- Gillespie, T.W.; Foody, G.M.; Rocchini, D.; Giorgi, A.P.; Saatchi, S. Measuring and modelling biodiversity from space. Prog. Phys. Geogr. 2008, 32, 203–221. [Google Scholar] [CrossRef] [Green Version]
- Skidmore, A.K.; Pettorelli, N. Agree on biodiversity metrics to track from space: Ecologists and space agencies must forge a global monitoring strategy. Nature 2015, 523, 403–406. [Google Scholar] [CrossRef] [PubMed]
- Hartemink, N.; Vanwambeke, S.O.; Purse, B.V.; Gilbert, M.; Van Dyck, H. Towards a resource-based habitat approach for spatial modelling of vector-borne disease risks. Biol. Rev. 2015, 90, 1151–1162. [Google Scholar] [CrossRef] [PubMed]
- Hutchinson, G.E. Concluding remarks. Cold Spring Harbor Symp. Quant. Biol. 1957, 22, 415–427. [Google Scholar] [CrossRef]
- Dennis, R.L.; Shreeve, T.G.; Van Dyck, H. Towards a functional resource-based concept for habitat: A butterfly biology viewpoint. Oikos 2003, 417–426. [Google Scholar]
- Jones, E.O.; Webb, S.D.; Ruiz-Fons, F.J.; Albon, S.; Gilbert, L. The effect of landscape heterogeneity and host movement on a tick-borne pathogen. Theor. Ecol. 2011, 4, 435–448. [Google Scholar] [CrossRef]
- Fahrig, L.; Baudry, J.; Brotons, L.; Burel, F.G.; Crist, T.O.; Fuller, R.J.; Sirami, C.; Siriwardena, G.M.; Martin, J.L. Functional landscape heterogeneity and animal biodiversity in agricultural landscapes. Ecol. Lett. 2011, 14, 101–112. [Google Scholar] [CrossRef]
- Li, S.; Hartemink, N.; Speybroeck, N.; Vanwambeke, S.O. Consequences of landscape fragmentation on Lyme disease risk: A cellular automata approach. PLoS ONE 2012, 7, e39612. [Google Scholar] [CrossRef]
- Li, S.; Heyman, P.; Cochez, C.; Simons, L.; Vanwambeke, S.O. A multi-level analysis of the relationship between environmental factors and questing Ixodes ricinus dynamics in Belgium. Parasites Vector 2012, 5, 149. [Google Scholar] [CrossRef]
- De Keukeleire, M.; Vanwambeke, S.O.; Somassè, E.; Kabamba, B.; Luyasu, V.; Robert, A. Scouts, forests, and ticks: Impact of landscapes on human-tick contacts. Ticks Tick Borne Dis. 2015, 6, 636–644. [Google Scholar] [CrossRef] [PubMed]
- Nupp, T.E.; Swihart, R.K. Effects of forest fragmentation on population attributes of white footed mice and eastern chipmunks. J. Mammol. 1998, 79, 1234–1243. [Google Scholar] [CrossRef]
- Allan, B.F.; Keesing, F.; Ostfeld, R.S. Effect of forest fragmentation on Lyme disease risk. Conserv. Biol. 2003, 17, 267–272. [Google Scholar] [CrossRef]
- Anderson, C.S.; Meikle, D.B. Annual changes in structural complexity of understory vegetation and relative abundance of Peromyscus leucopus in fragmented habitats. Acta Theriol. 2006, 51, 43–51. [Google Scholar] [CrossRef]
- Killilea, M.E.; Swei, A.; Lane, R.S.; Briggs, C.J.; Ostfeld, R.S. Spatial dynamics of Lyme disease: A review. EcoHealth 2008, 5, 167–195. [Google Scholar] [CrossRef]
- Gilbert, L.; Maffey, G.L.; Ramsay, S.L.; Hester, A.J. The effect of deer management on the abundance of Ixodes ricinus in Scotland. Ecol. Appl. 2012, 22, 658–667. [Google Scholar] [CrossRef]
- Gilbert, L. How landscapes shape Lyme borreliosis risk. In Ecology and Prevention of Lyme Borreliosis; Braks, M.A., van Wieren, S.E., Takken, W., Sprong, H., Eds.; Wageningen Academic Publishers: Wageningen, The Netherlands, 2016. [Google Scholar]
- Rocchini, D.; Bacaro, G.; Chirici, G.; Da Re, D.; Feilhauer, H.; Foody, G.M.; Gillespie, T.W.; He, K.S.; Kark, S.; Levin, N.; et al. Remotely sensed spatial heterogeneity as an exploratory tool for taxonomic and functional diversity study. Ecol. Indic. 2018, 85, 983–990. [Google Scholar] [CrossRef] [Green Version]
- Rocchini, D.; Delucchi, L.; Bacaro, G.; Cavallini, P.; Feilhauer, H.; Foody, G.M.; He, K.S.; Nagendra, H.; Porta, C.; Ricotta, C.; et al. Calculating landscape diversity with information-theory based indices: A GRASS GIS solution. Ecol. Inform. 2013, 17, 82–93. [Google Scholar] [CrossRef]
- De Clercq, E.M.; Vanwambeke, S.O. Classification of anthropogenic landscapes. In Analysis of Multitemporal Remote Sensing Images (MultiTemp). In Proceedings of the 9th International Workshop on the Analysis of Multitemporal Remote Sensing Images (MultiTemp), Brugge, Belgium, 27–29 June 2017. [Google Scholar] [CrossRef]
- Cherrill, A.; McClean, C. Between-observer variation in the application of a standard method of habitat mapping by environmental consultants in the UK. J. App. Ecol. 1999, 36, 989–1008. [Google Scholar] [CrossRef]
- Hearn, S.M.; Healey, J.R.; McDonald, M.A.; Turner, A.J.; Wong, J.L.G.; Stewart, G.B. The repeatability of vegetation classification and mapping. J. Environ. Manag. 2011, 92, 1174–1184. [Google Scholar] [CrossRef]
- Amici, V.; Filibeck, G.; Rocchini, D.; Geri, F.; Landi, S.; Giorgini, D.; Scoppola, A.; Chiarucci, A. Are CORINE land cover classes reliable proxies of plant species assemblages? A test in Mediterranean forest landscapes. Plant Biosyst. 2017, 152, 994–1001. [Google Scholar] [CrossRef]
- Guidigan, M.L.G.; Azihou, F.; Idohou, R.; Okhimamhe, A.A.; Fandohan, A.B.; Sinsin, B.; Adet, L. Modelling the current and future distribution of Kigelia africana under climate change in Benin, West Africa. Model. Earth Syst. Environ. 2018, 4, 1225–1238. [Google Scholar] [CrossRef]
- De Clercq, E.M.; Vanwambeke, S.O.; Sungirai, M.; Adehan, S.; Lokossou, R.; Madder, M. Geographic distribution of the invasive cattle tick Rhipicephalus microplus, a country-wide survey in Benin. Exp. Appl. Acarol. 2012, 58, 441–452. [Google Scholar] [CrossRef] [Green Version]
- De Clercq, E.M.; Leta, S.; Estrada-Peña, A.; Madder, M.; Adehan, S.; Vanwambeke, S.O. Species distribution modelling for Rhipicephalus microplus (Acari: Ixodidae) in Benin, West Africa: Comparing datasets and modelling algorithms. Prev. Vet. Med. 2015, 118, 8–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vancutsem, C.; Marinho, E.; Kayitakire, F.; See, L.; Fritz, S. Harmonizing and combining existing land cover/land use datasets for cropland area monitoring at the African continental scale. Remote Sens. 2012, 5, 19–41. [Google Scholar] [CrossRef]
- Delrue, J.; Bydekerke, L.; Eerens, H.; Gilliams, S.; Piccard, I.; Swinnen, E. Crop mapping in countries with small-scale farming: A case study for West Shewa, Ethiopia. Int. J. Remote Sens. 2013, 34, 2566–2582. [Google Scholar] [CrossRef]
- Lambert, M.J.; Waldner, F.; Defourny, P. Cropland mapping over Sahelian and Sudanian agrosystems: A knowledge-based approach using PROBA-V time series at 100-m. Remote Sens. 2016, 8, 232. [Google Scholar] [CrossRef]
- Walker, A.R. Ticks of Domestic Animals in Africa: A Guide to Identification of Species; Bioscience Reports; Bioscience: Edinburgh, UK, 2003; pp. 3–210. [Google Scholar]
- Madder, M.; Adehan, S.; De Deken, R.; Adehan, R.; Lokossou, R. New foci of Rhipicephalus microplus in West Africa. Exp. Appl. Acarol. 2012, 56, 385–390. [Google Scholar] [CrossRef] [Green Version]
- Madder, M.; Walker, J.G.; Van Rooyen, J.; Knobel, D.; De Clercq, E.M.; Vanwambeke, S.O.; Berkvens, D. e-Surveillance in animal health: Use and evaluation of mobile tools. Parasitology 2012, 139, 1831–1842. [Google Scholar] [CrossRef]
- Wallménius, K.; Barboutis, C.; Fransson, T.; Jaenson, T.G.; Lindgren, P.E.; Nyström, F.; Olsen, B.; Salaneck, E.; Nilsson, K. Spotted fever Rickettsia species in Hyalomma and Ixodes ticks infesting migratory birds in the European Mediterranean area. Parasit. Vectors 2014, 7, 318. [Google Scholar] [CrossRef]
- Sands, A.F.; Apanaskevich, D.A.; Matthee, S.; Horak, I.G.; Harrison, A.; Karim, S.; Mohammad, M.K.; Mumcuouglu, K.Y.; Rajakaruna, R.S.; Santos-Silva, M.M.; et al. Effects of tectonics and large scale climatic changes on the evolutionary history of Hyalomma ticks. Mol. Phylogenet. Evol. 2017, 114, 153–165. [Google Scholar] [CrossRef]
- Hill, M.J. Vegetation index suites as indicators of vegetation state in grassland and savanna: An analysis with simulated SENTINEL 2 data for a North American transect. Remote Sens. Environ. 2013, 137, 94–111. [Google Scholar] [CrossRef]
- Estrada-Peña, A.; Gray, J.S.; Kahl, O.; Lane, R.S.; Nijhoff, A.M. Research on the ecology of ticks and tick-borne pathogens—methodological principles and caveats. Cell. Infect. Microbiol. 2013, 3, 29. [Google Scholar] [CrossRef]
- Estrada-Peña, A.; Alexander, N.; Wint, G.W. Perspectives on modelling the distribution of ticks for large areas: So far so good? Parasit. Vectors 2016, 9, 179. [Google Scholar] [CrossRef]
- Rocchini, D.; Balkenhol, N.; Carter, G.A.; Foody, G.M.; Gillespie, T.W.; He, K.S.; Kark, S.; Levin, N.; Lucas, K.; Levin, N.; et al. Remotely sensed spectral heterogeneity as a proxy of species diversity: Recent advances and open challenges. Ecol. Inform. 2010, 5, 318–329. [Google Scholar] [CrossRef]
- Hernández-Stefanoni, J.L.; Gallardo-Cruz, J.A.; Meave, J.A.; Rocchini, D.; Bello-Pineda, J.; López-Martínez, J.O. Modeling α-and β-diversity in a tropical forest from remotely sensed and spatial data. Int. J. Appl. Earth Obs. 2012, 19, 359–368. [Google Scholar] [CrossRef]
- Rocchini, D.; Marcantonio, M.; Ricotta, C. Measuring Rao’s Q diversity index from remote sensing: An open source solution. Ecol. Indic. 2017, 72, 234–238. [Google Scholar] [CrossRef]
- Wallis, C.I.; Brehm, G.; Donoso, D.A.; Fiedler, K.; Homeier, J.; Paulsch, D.; Süßenbach, D.; Tiede, Y.; Brandl, R.; Farwig, N.; et al. Remote sensing improves prediction of tropical montane species diversity but performance differs among taxa. Ecol. Indic. 2017, 83, 538–549. [Google Scholar] [CrossRef]
- Haralick, R.M.; Shanmugam, K. Textural features for image classification. IEEE Trans. Syst. Man. Cybern. 1973, 3, 610–621. [Google Scholar] [CrossRef]
- Zhang, X.; Cui, J.; Wang, W.; Lin, C. A study for texture feature extraction of high-resolution satellite images based on a direction measure and gray level co-occurrence matrix fusion algorithm. Sensors 2017, 17, 1474. [Google Scholar] [CrossRef]
- Wood, E.M.; Pidgeon, A.M.; Radeloff, V.C.; Keuler, N.S. Image texture as a remotely sensed measure of vegetation structure. Remote Sens. Environ. 2012, 121, 516–526. [Google Scholar] [CrossRef]
- Wood, E.M.; Pidgeon, A.M.; Radeloff, V.C.; Keuler, N.S. Image texture predicts avian density and species richness. PLoS ONE 2013, 8, e63211. [Google Scholar] [CrossRef] [PubMed]
- Arino, O.; Ramos Perez, J.J.; Kalogirou, V.; Bontemps, S.; Defourny, P.; Van Bogaert, E. Global land cover map for 2009 (GlobCover 2009). ESA & UCL. 2012. Available online: http://due.esrin.esa.int/page_globcover.php (accessed on February 2018).
- Lambert, D. Zero-inflated Poisson regression, with an application to defects in manufacturing. Technometrics 1992, 34, 1–14. [Google Scholar] [CrossRef]
- Vuong, Q.H. Likelihood ratio tests for model selection and non-nested hypotheses. Econometrica 1989, 307–333. [Google Scholar] [CrossRef]
- Dormann, C.F.; Elith, J.; Bacher, S.; Buchmann, C.; Carl, G.; Carré, G.; Garcia Marquéz, J.R.; Gruber, B.; Lafourcade, B.; Leitão, P.J.; et al. Collinearity: A review of methods to deal with it and a simulation study evaluating their performance. Ecography 2013, 36, 27–46. [Google Scholar] [CrossRef]
- Burnham, K.P.; Anderson, D.R. Model Selection and Multimodel Inference: A Practical Information-Theoretic Approach; Springer: New York, NY, USA, 2002. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019; Available online: http://www.R-project.org/ (accessed on 1 February 2019).
- Rocchini, D.; Ricotta, C.; Chiarucci, A.; De Dominicis, V.; Cirillo, I.; Maccherini, S. Relating spectral and species diversity through rarefaction curves. Int. J. Remote Sens. 2009, 30, 2705–2711. [Google Scholar] [CrossRef]
- Rocchini, D.; Neteler, M. Spectral rank–abundance for measuring landscape diversity. Int. J. Remote Sens. 2012, 33, 4458–4470. [Google Scholar] [CrossRef] [Green Version]
- Randolph, S.E. Ticks and tick-borne disease systems in space and from space. Adv. Parasitol. 2000, 47, 217–243. [Google Scholar] [CrossRef]
- Warren, S.D.; Alt, M.; Olson, K.D.; Irl, S.D.; Steinbauer, M.J.; Jentsch, A. The relationship between the spectral diversity of satellite imagery, habitat heterogeneity, and plant species richness. Ecol. Inform. 2014, 24, 160–168. [Google Scholar] [CrossRef]
- Marino, P.C.; Landis, D.A. Effect of landscape structure onparasitoid diversity and parasitism in agroecosystems. Ecol. Appl. 1996, 6, 276–284. [Google Scholar] [CrossRef]
- Guerrini, L.; Bord, J.P.; Ducheyne, E.; Bouyer, J. Fragmentation analysis for prediction of suitable habitat for vectors: Example of riverine tsetse flies in Burkina Faso. J. Med. Entomol. 2008, 45, 1180–1186. [Google Scholar] [CrossRef] [PubMed]
- Gardiner, M.; Landis, D.; Gratton, C.; DiFonzo, C.; O’neal, M.; Chacon, J.M.; Wayo, M.T.; Schmidt, N.P.; Mueller, E.E.; Heimpel, G.E. Landscape diversity enhances biological control of an introduced crop pest in the north-central USA. Ecol. Appl. 2009, 19, 143–154. [Google Scholar] [CrossRef] [Green Version]
- Penn, H.J.; Athey, K.J.; Lee, B.D. Land cover diversity increases predator aggregation and consumption of prey. Ecol. Lett. 2017, 20, 609–618. [Google Scholar] [CrossRef]
- Hofmann, S.; Everaars, J.; Schweiger, O.; Frenzel, M.; Bannehr, L.; Cord, A.F. Modelling patterns of pollinator species richness and diversity using satellite image texture. PLoS ONE 2017, 12, e0185591. [Google Scholar] [CrossRef] [PubMed]
- Apanaskevich, D.A.; Schuster, A.L.; Horak, I.G. The genus Hyalomma: VII. Redescription of all parasitic stages of H.(Euhyalomma) dromedarii and H.(E.) schulzei (Acari: Ixodidae). J. Med. Entomol. 2008, 45, 817–831. [Google Scholar] [CrossRef]
- Guglielmone, A.A.; Robbins, R.G.; Apanaskevich, D.A.; Petney, T.N.; Estrada-Peña, A.; Horak, I.G. The Hard Ticks of the World; Springer: Dordrecht, The Netherlands, 2014; Volume 10, pp. 978–994. [Google Scholar]
- Miguel, E.; Boulinier, T.; de Garine-Wichatitsky, M.; Caron, A.; Fritz, H.; Grosbois, V. Characterising African tick communities at a wild–domestic interface using repeated sampling protocols and models. Acta Trop. 2014, 138, 5–14. [Google Scholar] [CrossRef] [Green Version]
- Spengler, J.R.; Estrada-Peña, A. Host preferences support the prominent role of Hyalomma ticks in the ecology of Crimean-Congo hemorrhagic fever. PLoS Negl. Trop. Dis. 2018, 12, e0006248. [Google Scholar] [CrossRef]
- Vanwambeke, S.O.; Li, S.; Hartemink, N.A. A resource-based habitat concept for tick-borne diseases. In Ecology and Prevention of Lyme Borreliosis; Braks, M.A., van Wieren, S.E., Takken, W., Sprong, H., Eds.; Wageningen Academic Publishers: Wageningen, The Netherlands, 2016; pp. 205–216. [Google Scholar]
- England, M. Understanding the Risks and Factors Associated with the Introduction of Crimean-Congo Haemorrahagic Fever Virus into Great Britain. Ph.D. Dissertation, University of Southampton, Southampton, UK, 2015. [Google Scholar]
- Gilot, B.; Guiguen, C.; Degeilh, B.; Doche, B.; Pichot, J.; Beaucournu, J.C. Phytoecological mapping of Ixodes ricinus as an approach to the distribution of Lyme borreliosis in France. In Lyme Borreliosis; Axford, J.S., Rees, D.H.E., Eds.; NATO ASI Series (Series A: Life Sciences); Springer: Boston, MA, USA, 1994; Volume 260, pp. 105–112. [Google Scholar]
- Karlson, M.; Ostwald, M.; Reese, H.; Sanou, J.; Tankoano, B.; Mattsson, E. Mapping TreeCanopy Cover and Aboveground Biomass in Sudano-Sahelian Woodlands Using Landsat 8 and Random Forest. Remote Sens. 2015, 7, 10017–10041. [Google Scholar] [CrossRef]
- Godinho, S.; Guiomar, N.; Gil, A. Estimating tree canopy cover percentage in a mediterranean silvopastoral systems using Sentinel-2A imagery and the stochastic gradient boosting algorithm. Int. J. Remote Sens. 2018, 39, 4640–4662. [Google Scholar] [CrossRef]
- Rocchini, D. Effects of spatial and spectral resolution in estimating ecosystem α-diversity by satellite imagery. Remot. Sen. Environ. 2007, 111, 423–434. [Google Scholar] [CrossRef]





| Predictors | Abbreviation |
|---|---|
| Spectral predictors | |
| Multispectral Rao’s Q | rao_ms |
| Rao’s Q PC1 | rao_pc1 |
| GLCM PC1 variance | pc1_var |
| GLCM PC1 entropy | pc1_entr |
| GLCM PC1 contrast | pc1_contr |
| NDVI | |
| NDVI | ndvi |
| Rao’s Q NDVI | rao_ndvi |
| GLCM NDVI variance | ndvi_var |
| GLCM NDVI entropy | ndvi_entr |
| GLCM NDVI contrast | ndvi_contr |
| Value | Label | % |
|---|---|---|
| 14 | Rainfed croplands | 2.59 |
| 20 | Mosaic cropland (50–70%)/vegetation (grassland/shrubland/forest) (20–50%) | 28.54 |
| 30 | Mosaic vegetation (grassland/shrubland/forest) (50–70%)/cropland (20–50%) | 10.95 |
| 40 | Closed to open (>15%) broadleaved evergreen or semi-deciduous forest (>5 m) | 1.44 |
| 60 | Open (15–40%) broadleaved deciduous forest/woodland (>5 m) | 9.67 |
| 110 | Mosaic forest or shrubland (50–70%)/grassland (20–50%) | 2.68 |
| 120 | Mosaic grassland (50–70%)/forest or shrubland (20–50%) | 0.29 |
| 130 | Closed to open (>15%) (broadleaved or needleleaved, evergreen or deciduous) shrubland (<5 m) | 43.71 |
| 140 | Closed to open (>15%) herbaceous vegetation (grassland, savannas or lichens/mosses) | 2.68 |
| Spectral Predictor | NDVI Predictors | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Predictors | Count model | ZI Binomial | AIC | Predictors | Count model | ZI Binomial | AIC | ||||||||||||
| Estimate | 2.5% CI | 97.5% CI | p.val | Estimate | 2.5% CI | 97.5% CI | p.val | Estimate | 2.5% CI | 97.5% CI | p.val | Estimate | 2.5% CI | 97.5% CI | p.val | ||||
| Intercept | 16.32 | 15.51 | 17.17 | *** | 0.14 | 0.08 | 0.26 | *** | 2454.71 | - | - | - | - | - | - | - | - | - | - |
| Intercept | 16.19 | 15.38 | 17.04 | *** | 0.13 | 0.07 | 0.25 | *** | 2403.68 | Intercept | 16.32 | 15.51 | 17.17 | *** | 0.14 | 0.08 | 0.25 | *** | 2458.56 |
| Multispectral Rao’s Q | 0.83 | 0.78 | 0.87 | *** | 0.61 | 0.31 | 1.18 | ns | NDVI | 1.00 | 0.95 | 1.05 | ns | 1.12 | 0.63 | 2.00 | ns | ||
| Intercept | 16.28 | 15.47 | 17.13 | *** | 0.13 | 0.07 | 0.25 | *** | 2432.81 | Intercept | 15.36 | 14.55 | 16.22 | *** | 0.14 | 0.08 | 0.26 | *** | 2303.15 |
| Rao’s Q PC1 | 0.88 | 0.84 | 0.93 | *** | 0.67 | 0.35 | 1.28 | ns | Rao’s Q NDVI | 0.68 | 0.63 | 0.72 | *** | 0.95 | 0.52 | 1.74 | ns | ||
| Intercept | 16.32 | 15.51 | 17.17 | *** | 0.12 | 0.06 | 0.24 | *** | 2454.96 | Intercept | 16.32 | 15.51 | 17.17 | *** | 0.14 | 0.08 | 0.26 | *** | 2458.53 |
| GLCM PC1 variance | 1.00 | 0.95 | 1.05 | ns | 1.95 | 0.93 | 4.10 | ns | GLCM NDVI variance | 0.99 | 0.95 | 1.04 | ns | 1.10 | 0.62 | 1.97 | ns | ||
| Intercept | 16.32 | 15.51 | 17.17 | *** | 0.14 | 0.08 | 0.25 | *** | 2450.14 | Intercept | 15.97 | 15.16 | 16.82 | *** | 0.14 | 0.08 | 0.25 | *** | 2373.31 |
| GLCM PC1 entropy | 0.93 | 0.89 | 0.98 | ** | 0.74 | 0.42 | 1.30 | ns | GLCM NDVI entropy | 0.78 | 0.73 | 0.82 | *** | 0.78 | 0.42 | 1.43 | ns | ||
| Intercept | 16.25 | 15.44 | 17.10 | *** | 0.13 | 0.07 | 0.25 | *** | 2422.28 | Intercept | 15.96 | 15.15 | 16.82 | *** | 0.14 | 0.08 | 0.25 | ns | 2382.06 |
| GLCM PC1 contrast | 0.85 | 0.81 | 0.90 | *** | 0.57 | 0.25 | 1.28 | ns | GLCM NDVI contrast | 0.77 | 0.72 | 0.82 | *** | 0.74 | 0.35 | 1.54 | *** | ||
| Spectral Predictors | NDVI Predictors | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Predictors | Count Model | ZI Binomial | AIC | Predictors | Count model | ZI Binomial | AIC | ||||||||||||
| Estimate | 2.5% CI | 97.5% CI | p.val | Estimate | 2.5% CI | 97.5% CI | p.val | Estimate | 2.5% CI | 97.5% CI | p.val | Estimate | 2.5% CI | 97.5% CI | p.val | ||||
| Intercept | 14.96 | 13.87 | 16.13 | *** | 1.31 | 0.89 | 1.93 | ns | 785.47 | - | - | - | - | - | - | - | - | - | - |
| Intercept | 14.39 | 13.22 | 15.66 | *** | 1.35 | 0.88 | 2.05 | ns | 767.84 | Intercept | 14.02 | 12.77 | 15.39 | *** | 1.45 | 0.92 | 2.27 | ns | 757.86 |
| Multispectral Rao’s Q | 1.08 | 1.01 | 1.16 | * | 0.39 | 0.24 | 0.65 | *** | NDVI | 0.89 | 0.82 | 0.98 | * | 3.27 | 1.91 | 5.61 | *** | ||
| Intercept | 14.41 | 13.21 | 15.71 | *** | 1.35 | 0.88 | 2.08 | ns | 765.24 | Intercept | 13.59 | 12.43 | 14.87 | *** | 1.36 | 0.91 | 2.05 | ns | 760.05 |
| Rao’s Q PC1 | 1.07 | 1.00 | 1.16 | ns | 0.35 | 0.21 | 0.59 | *** | Rao’s Q NDVI | 0.76 | 0.67 | 0.85 | *** | 1.86 | 1.13 | 3.07 | * | ||
| Intercept | 12.70 | 11.50 | 14.02 | *** | 1.33 | 0.84 | 2.10 | *** | 724.21 | Intercept | 13.89 | 12.64 | 15.27 | *** | 1.47 | 0.93 | 2.30 | ns | 756.63 |
| GLCM PC1 variance | 0.79 | 0.73 | 0.86 | *** | 4.10 | 2.24 | 7.50 | ns | GLCM NDVI variance | 0.88 | 0.80 | 0.96 | ** | 3.28 | 1.90 | 5.66 | *** | ||
| Intercept | 13.73 | 12.47 | 15.11 | *** | 1.46 | 0.94 | 2.28 | ns | 756.78 | Intercept | 14.75 | 13.60 | 15.99 | *** | 1.34 | 0.90 | 2.01 | ns | 781.49 |
| GLCM PC1 entropy | 1.17 | 1.06 | 1.29 | ** | 0.32 | 0.19 | 0.55 | *** | GLCM NDVI entropy | 0.96 | 0.87 | 1.04 | ns | 1.73 | 1.13 | 2.66 | * | ||
| Intercept | 14.69 | 13.51 | 15.96 | *** | 1.28 | 0.83 | 1.97 | ns | 767.07 | Intercept | 14.65 | 13.53 | 15.87 | *** | 1.34 | 0.90 | 1.99 | ns | 782.91 |
| GLCM PC1 contrast | 1.04 | 0.97 | 1.11 | ns | 0.33 | 0.19 | 0.58 | *** | GLCM NDVI contrast | 0.91 | 0.83 | 1.01 | ns | 1.51 | 0.94 | 2.42 | ns | ||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Da Re, D.; De Clercq, E.M.; Tordoni, E.; Madder, M.; Rousseau, R.; Vanwambeke, S.O. Looking for Ticks from Space: Using Remotely Sensed Spectral Diversity to Assess Amblyomma and Hyalomma Tick Abundance. Remote Sens. 2019, 11, 770. https://doi.org/10.3390/rs11070770
Da Re D, De Clercq EM, Tordoni E, Madder M, Rousseau R, Vanwambeke SO. Looking for Ticks from Space: Using Remotely Sensed Spectral Diversity to Assess Amblyomma and Hyalomma Tick Abundance. Remote Sensing. 2019; 11(7):770. https://doi.org/10.3390/rs11070770
Chicago/Turabian StyleDa Re, Daniele, Eva M. De Clercq, Enrico Tordoni, Maxime Madder, Raphaël Rousseau, and Sophie O. Vanwambeke. 2019. "Looking for Ticks from Space: Using Remotely Sensed Spectral Diversity to Assess Amblyomma and Hyalomma Tick Abundance" Remote Sensing 11, no. 7: 770. https://doi.org/10.3390/rs11070770
APA StyleDa Re, D., De Clercq, E. M., Tordoni, E., Madder, M., Rousseau, R., & Vanwambeke, S. O. (2019). Looking for Ticks from Space: Using Remotely Sensed Spectral Diversity to Assess Amblyomma and Hyalomma Tick Abundance. Remote Sensing, 11(7), 770. https://doi.org/10.3390/rs11070770