Retrieving Phytoplankton Size Class from the Absorption Coefficient and Chlorophyll A Concentration Based on Support Vector Machine
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Sampling and Optical Measurements
2.3. Phytoplankton Pigment-Based Size Classes
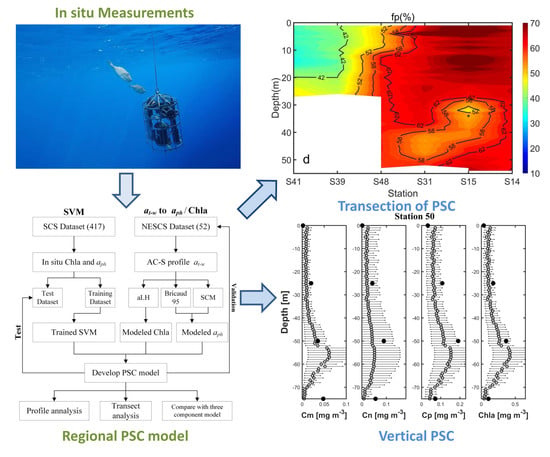
2.4. Reconstruction of aph and Chla from at-w
2.5. Development of Vertical PSC Model
2.6. Assessments
3. Results
3.1. Distribution of PSCs
3.2. Selection of Input Parameters
3.3. Cross-Validation Tests
3.4. Results of the PSC Model
3.5. Preliminary Application of Transect Distribution
3.6. Comparisons with the Three-Component Model
4. Discussion
4.1. Errors Introduced Via Reconstruction of Chla Using aLH Methods Instead of Measurement
4.2. Errors from the Reconstruction of aph(λ)
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Falkowski, P.G.; Katz, M.E.; Knoll, A.H.; Quigg, A.; Raven, J.A.; Schofield, O.; Taylor, F.J. The evolution of modern eukaryotic phytoplankton. Science 2004, 305, 354–360. [Google Scholar] [CrossRef] [PubMed]
- Field, C.; Behrenfeld, M.; Randerson, J.; Falkowsk, P. Primary Production of the Biosphere: Integrating Terrestrial and Oceanic Components. Science 1998, 281, 237–240. [Google Scholar] [CrossRef]
- Longhurst, A.; Sathyendranath, S.; Platt, T.; Caverhill, C. An estimate of global primary production in the ocean from satellite radiometer data. J. Plank. Res. 1995, 17, 1245–1271. [Google Scholar] [CrossRef]
- Le Quéré, C.; Harrison, S.P.; Prentice, C.I.; Buitenhuis, E.T.; Aumont, O.; Bopp, L.; Claustre, H. Ecosystem dynamics based on plankton functional types for global ocean biogeochemistry models. Glob. Change Bio. 2005, 11, 2016–2040. [Google Scholar] [CrossRef]
- Geider, R.J.; Platt, T.; Raven, J.A. Size dependence of growth and photosynthesis in diatoms: A synthesis. Mar. Ecol. Prog. Ser. 1986, 30, 93–104. [Google Scholar] [CrossRef]
- Maloney, C.L.; Field, J.G. The size-based dynamics of plankton food webs. I. A simulation model of carbon and nitrogen flows. J. Plank. Res. 1991, 13, 1003–1038. [Google Scholar] [CrossRef]
- Parsons, T.R.; Lalli, C.M. Jellyfish populations explosions: Revisiting a hypothesis of possible causes. La Mer 2002, 40, 640–647. [Google Scholar]
- Platt, T.; Denman, K.L. The relationship between photosynthesis and light for natural assemblages of coastal marine phytoplankton. J. Phyco. 1976, 12, 421–430. [Google Scholar] [CrossRef]
- Probyn, T.A. Nitrogen uptake by size-fractionated phytoplankton populations in the southern Benguela upwelling system. Mar. Ecol. Prog. Ser. 1985, 22, 249–258. [Google Scholar] [CrossRef]
- Sieburth, J.; Smetacek, V.; Lenz, J. Pelagic ecosystem structure: Heterotrophic compartments of the plankton and their relationship to plankton size fraction. Limnol. Oceanogr. 1978, 23, 1256–1263. [Google Scholar] [CrossRef]
- IOCCG. Phytoplankton Functional Types from Space. In Reports of the International Ocean-Colour Coordinating Group; Sathyendranath, S., Ed.; IOCCG: Dartmouth, NS, Canada, 2014. [Google Scholar]
- Brewin, R.; Sathyendranath, S.; Hirata, T.; Lavender, S.J.; Barciela, R.M.; Hardman-Mountford, N.J. A three-component model of phytoplankton size class for the Atlantic Ocean. Ecol. Model. 2010, 221, 1472–1483. [Google Scholar] [CrossRef]
- Hirata, T.; Hardman-Mountford, N.J.; Brewin, R.J.W.; Aiken, J.; Barlow, R.; Suzuki, K.; Isada, T.; Howell, E.; Hashioka, T.; Noguchi-Aita, M.; et al. Synoptic relationships between surface Chlorophyll-a and diagnostic pigments specific to phytoplankton functional types. Biogeosci. 2011, 8, 311–327. [Google Scholar] [CrossRef]
- Sathyendranath, S.; Cota, G.; Stuart, V.; Maass, H.; Platt, T. Remote sensing of phytoplankton pigments: A comparison of empirical and theoretical approaches. Int. J. Remote Sens. 2001, 22, 249–273. [Google Scholar] [CrossRef]
- Uitz, J.; Claustre, H.; Morel, A.; Hooker, S.B. Vertical distribution of phytoplankton communities in open ocean: An assessment based on surface chlorophyll. J. Geophys. Res. 2006, 111. [Google Scholar] [CrossRef]
- Ciotti, A.; Bricaud, A. Retrievals of a size parameter for phytoplankton and spectral light absorption by coloured detrital matter from water-leaving radiances at SeaWiFS channels in a continental shelf off Brazil. Limnol. Oceanogr. 2006, 4, 237–253. [Google Scholar] [CrossRef]
- Ciotti, A.; Lewis, M.R.; Cullen, J.J. Assessment of the Relationships between Dominant Cell Size in Natural Phytoplankton Communities and the Spectral Shape of the Absorption Coefficient. Limnol. Oceanogr. 2002, 47, 404–417. [Google Scholar] [CrossRef]
- Devred, E.; Sathyendranath, S.; Stuart, V.; Maass, H.; Ulloa, O.; Platt, T. A two-component model of phytoplankton absorption in the open ocean: Theory and applications. J. Geophys. Res. 2006, 111. [Google Scholar] [CrossRef]
- Hirata, T.; Aiken, J.; Hardman-Mountford, N.; Smyth, T.J.; Barlow, R.G. An absorption model to determine phytoplankton size classes from satellite ocean colour. Remote Sens. Environ. 2008, 112, 3153–3159. [Google Scholar] [CrossRef]
- Roy, S.; Sathyendranath, S.; Bouman, H.; Platt, T. The global distribution of phytoplankton size spectrum and size classes from their light-absorption spectra derived from satellite data. Remote Sens. Environ. 2013, 139, 185–197. [Google Scholar] [CrossRef]
- Kostadinov, T.S.; Siegel, D.A.; Maritorena, S. Retrieval of the particle size distribution from satellite ocean color observations. J. Geophys. Res. 2009, 114, 1–22. [Google Scholar] [CrossRef]
- Emanuele, O.; Annick, B.; David, A.; Julia, U. Multivariate approach for the retrieval of phytoplankton size structure from measured light absorption spectra in the Mediterranean Sea. Appl. Opt. 2013, 52, 2257–2273. [Google Scholar]
- Wang, S.; Ishizaka, J.; Hirawake, T.; Watanabe, Y.; Zhu, Y.; Hayashi, M.; Yoo, S. Remote estimation of phytoplankton size fractions using the spectral shape of light absorption. Opt. Express 2015, 23, 10301–10318. [Google Scholar] [CrossRef]
- Catlett, D.; Siegel, D.A. Phytoplankton Pigment Communities Can be Modeled Using Unique Relationships with Spectral Absorption Signatures in a Dynamic Coastal Environment. J. Geophys. Res. 2018, 123, 246–264. [Google Scholar] [CrossRef]
- Torrecilla, E.; Stramski, D.; Reynolds, R.A.; Millán-Núñez, E.; Piera, J. Cluster analysis of hyperspectral optical data for discriminating phytoplankton pigment assemblages in the open ocean. Remote Sens. Environ. 2011, 115, 2578–2593. [Google Scholar] [CrossRef]
- Uitz, J.; Stramski, D.; Reynolds, R.A.; Dubranna, J. Assessing phytoplankton community composition from hyperspectral measurements of phytoplankton absorption coefficient and remote-sensing reflectance in open-ocean environments. Remote Sens. Environ. 2015, 171, 58–74. [Google Scholar] [CrossRef]
- Brewin, R.J.W.; Hardman-Mountford, N.J.; Lavender, S.J.; Raitsos, D.E.; Hirata, T.; Uitz, J.; Devred, E.; Bricaud, A.; Ciotti, A.; Gentili, B. An intercomparison of bio-optical techniques for detecting dominant phytoplankton size class from satellite remote sensing. Remote Sens. Environ. 2011, 115, 325–339. [Google Scholar] [CrossRef]
- Raitsos, D.E.; Lavender, S.J.; Maravelias, C.D.; Haralambous, J.; Richardson, A.J.; Reid, P.C. Identifying phytoplankton functional groups from space: An ecological approach. Limnol. Oceanogr. 2008, 53, 605–613. [Google Scholar] [CrossRef]
- Li, Z.; Li, L.; Song, K.; Cassar, N. Estimation of phytoplankton size fractions based on spectral features of remote sensing ocean color data. J. Geophys. Res. 2013, 118, 1445–1458. [Google Scholar] [CrossRef]
- Hu, S.; Liu, H.; Zhao, W.; Shi, T.; Hu, Z.; Li, Q.; Wu, G. Comparison of Machine Learning Techniques in Inferring Phytoplankton Size Classes. Remote Sens. 2018, 10. [Google Scholar] [CrossRef]
- Brewin, R.J.W.; Hirata, T.; Hardman-Mountford, N.J.; Lavender, S.J.; Sathyendranath, S.; Barlow, R. The influence of the Indian Ocean Dipole on interannual variations in phytoplankton size structure as revealed by Earth Observation. Deep Sea Res. II 2012, 77–80, 117–127. [Google Scholar] [CrossRef]
- Mouw, C.B.; Yoder, J.A. Optical determination of phytoplankton size composition from global SeaWiFS imagery. J. Geophys. Res. 2010, 115. [Google Scholar] [CrossRef]
- Nair, A.; Sathyendranath, S.; Platt, T.; Morales, J.; Stuart, V.; Forget, M.-H.; Devred, E.; Bouman, H. Remote sensing of phytoplankton functional types. Remote Sens. Environ. 2008, 112, 3366–3375. [Google Scholar] [CrossRef]
- Dai, M.; Zhai, W.; Cai, W.-J.; Callahan, J.; Huang, B.; Shang, S.; Huang, T.; Li, X.; Lu, Z.; Chen, W.; et al. Effects of an estuarine plume-associated bloom on the carbonate system in the lower reaches of the Pearl River estuary and the coastal zone of the northern South China Sea. Cont. Shelf Res. 2008, 28, 1416–1423. [Google Scholar] [CrossRef]
- Mitchell, B.G. Algorithms for determining the absorption coefficient of aquatic particulates using the quantitative filter technique (QFT). SPIE 1990, 1302, 137–148. [Google Scholar]
- Yentsch, C.S. Measurement of Visible Light Absorption by Particulate Matter in the Ocean. Limnol. Oceanogr. 1962, 7, 207–217. [Google Scholar] [CrossRef]
- Kisino, M.; Takahashi, M.; Okami, N.; Ichimura, S. Estimation of the spectral absorption coefficients of phytoplankton in the sea. Bull. Mar. Sci. 1985, 37, 634–642. [Google Scholar]
- Babin, M.; Stramski, D.; Ferrari, G.; Claustre, H.; Bricaud, A.; Obolensky, G.; Hoepffner, N. Variations in the light absorption coefficients of phytoplankton, nonalgal particles, and dissolved organic matter in coastal waters around Europe. J. Geophys. Res. 2003, 108. [Google Scholar] [CrossRef]
- Bricaud, A.; Morel, A.; Babin, M.; Allali, K.; Claustre, H. Variations of light absorption by suspended particles with chlorophyllaconcentration in oceanic (case 1) waters: Analysis and implications for bio-optical models. J. Geophys. Res. 1998, 103, 31033–31044. [Google Scholar] [CrossRef]
- Bricaud, A.; Stramski, D. Spectral absorption coefficients of living phytoplankton and nonalgal biogenous matter: A comparison between the Peru upwelling area and the Sargasso Sea. Limnol. Oceanogr. 1990, 35, 562–582. [Google Scholar] [CrossRef]
- Roesler, C.S. Theoretical and experimental approaches to improve the accuracy of particulate absorption coefficients from the Quantitative Filter Technique. Limnol. Oceanogr. 1998, 43, 11. [Google Scholar] [CrossRef]
- Vidussi, F.; Claustre, H.; Manca, B.B.; Luchetta, A.; Marty, J.-C. Phytoplankton pigment distribution in relation to upper thermocline circulation in the eastern Mediterranean Sea during winter. J. Geophys. Res. 2001, 106, 19939–19956. [Google Scholar] [CrossRef]
- Sullivan, J.; Twardowski, M.; Zaneveld, J.R.V.; Moore, C.; Barnard, A.H.; Donaghay, P.L.; Rhoades, B. Hyperspectral temperature and salt dependencies of absorption by water and heavy water in the 400–750 nm spectral range. Appl. Opt. 2006, 45, 5294–5309. [Google Scholar] [CrossRef]
- Zaneveld, J.R.V.; Kitchen, J.C.; Moore, C.C. Scattering error correction of reflecting-tube absorption meters. SPIE Ocean Opt. XII 1994, 2258, 44–55. [Google Scholar]
- Claustre, H. The Trophic Status of Various Oceanic Provinces as Revealed by Phytoplankton Pigment Signatures. Limnol. Oceanogr. 1994, 39, 1206–1210. [Google Scholar] [CrossRef]
- Roesler, C.S.; Barnard, A.H. Optical proxy for phytoplankton biomass in the absence of photophysiology: Rethinking the absorption line height. Methods in Oceanogr. 2013, 7, 79–94. [Google Scholar] [CrossRef]
- Bricaud, A.; Babin, M.; Morel, A.; Claustre, H. Variability in the chlorophyll-specific absorption coefficients of natural phytoplankton: Analysis and parameterization. J. Geophys. Res. 1995, 100, 13321. [Google Scholar] [CrossRef]
- Zheng, G.; Stramski, D. A model based on stacked-constraints approach for partitioning the light absorption coefficient of seawater into phytoplankton and non-phytoplankton components. J. Geophys. Res. 2013, 118, 2155–2174. [Google Scholar] [CrossRef]
- Zheng, G.; Stramski, D.; DiGiacomo, P.M. A model for partitioning the light absorption coefficient of natural waters into phytoplankton, nonalgal particulate, and colored dissolved organic components: A case study for the Chesapeake Bay. J. Geophys. Res. 2015, 120, 2601–2621. [Google Scholar] [CrossRef]
- Boss, E.S.; Collier, R.; Larson, G.; Fennel, K.; Pegau, W.S. Measurements of spectral optical properties and their relation to biogeochemical variables and processes in Crater Lake, Crater Lake National Park, OR. Hydrobiology 2007, 574, 149–159. [Google Scholar] [CrossRef]
- Boss, E.; Picheral, M.; Leeuw, T.; Chase, A.; Karsenti, E.; Gorsky, G.; Taylor, L.; Slade, W.; Ras, J.; Claustre, H. The characteristics of particulate absorption, scattering and attenuation coefficients in the surface ocean; Contribution of the Tara Oceans expedition. Methods Oceanogr. 2013, 7, 52–62. [Google Scholar] [CrossRef]
- Lin, J.; Cao, W.; Zhou, W.; Sun, Z.; Xu, Z.; Wang, G.; Hu, S. Novel method for quantifying the cell size of marine phytoplankton based on optical measurements. Opt. Express 2014, 22, 10467–10476. [Google Scholar] [CrossRef] [PubMed]
- Lamont, T.; Barlow, R.; Brewin, R. Variations in Remotely-Sensed Phytoplankton Size Structure of a Cyclonic Eddy in the Southwest Indian Ocean. Remote Sens. 2018, 10. [Google Scholar] [CrossRef]
- Sahay, A.; Ali, S.M.; Gupta, A.; Goes, J.I. Ocean color satellite determinations of phytoplankton size class in the Arabian Sea during the winter monsoon. Remote Sens. Environ. 2017, 198, 286–296. [Google Scholar] [CrossRef]
- Varunan, T.; Shanmugam, P. A model for estimating size-fractioned phytoplankton absorption coefficients in coastal and oceanic waters from satellite data. Remote Sens. Environ. 2015, 158, 235–254. [Google Scholar] [CrossRef]
- Hu, S.; Zhou, W.; Wang, G.; Cao, W.; Xu, Z.; Liu, H.; Wu, G.; Zhao, W. Comparison of Satellite-Derived Phytoplankton Size Classes Using In-Situ Measurements in the South China Sea. Remote Sens. 2018, 10. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, S.; Qiu, Z.; Sun, D.; Ishizaka, J.; Sun, S.; He, Y. Phytoplankton size class in the East China Sea derived from MODIS satellite data. Biogeoscience 2018, 15, 4271–4289. [Google Scholar] [CrossRef]
- Lin, J.; Cao, W.; Zhou, W.; Hu, S.; Wang, G.; Sun, Z.; Xu, Z.; Song, Q. A bio-optical inversion model to retrieve absorption contributions and phytoplankton size structure from total minus water spectral absorption using genetic algorithm. Chin. J. Ocean Limnol. 2013, 31, 970–978. [Google Scholar] [CrossRef]
- Huang, B.; Hu, J.; Xu, H.; Cao, Z.; Wang, D. Phytoplankton community at warm eddies in the northern South China Sea in winter 2003/2004. Deep Sea Res. II 2010, 57, 1792–1798. [Google Scholar] [CrossRef]
- Waga, H.; Hirawake, T.; Fujiwara, A.; Kikuchi, T.; Nishino, S.; Suzuki, K.; Takao, S.; Saitoh, S.-I. Differences in Rate and Direction of Shifts between Phytoplankton Size Structure and Sea Surface Temperature. Remote Sens. 2017, 9. [Google Scholar] [CrossRef]
- Chase, A.; Boss, E.; Zaneveld, R.; Bricaud, A.; Claustre, H.; Ras, J.; Dall’Olmo, G.; Westberry, T.K. Decomposition of in situ particulate absorption spectra. Methods Oceanogr. 2013, 7, 110–124. [Google Scholar] [CrossRef]
- Hoepffner, N.; Sathyendranath, S. Effect of pigment composition on absorption properties of phytoplankton. Mar. Ecol. Prog. Ser. 1991, 73, 11–23. [Google Scholar] [CrossRef]
- Hoepffner, N.; Sathyendranath, S. Determination of the major groups of phytoplankton pigments from the absorption spectra of total particulate matter. J. Geophys. Res. 1993, 98. [Google Scholar] [CrossRef]












| Data Type and Number of Points | |||||||
|---|---|---|---|---|---|---|---|
| Training | Test | Apply | Validate | ||||
| Expedition | Cruise Period | Locations | HPLC and aph(λ) | HPLC and aph(λ) | HPLC and at-w(λ) | at-w(λ) and aph(λ) | Totals |
| SCS | September 2006–August 2013 | South China Sea | 334 | 83 | 417 | ||
| NESCS | 5–17 August 2015 | Northeast South China Sea | 52 | 52 | |||
| WSCS | 9 August–2 September 2013 1–23 October 2017 | North South China Sea | 114 | 114 | |||
| Input Models | Size | Train | Test | ||||||
|---|---|---|---|---|---|---|---|---|---|
| RMS | APD% | RPD% | r2 | RMS | APD% | RPD% | r2 | ||
| SVM-Type1 | pico | 0.0496 | 15.15 | 4.26 | 0.88 | 0.0407 | 27.73 | 8.73 | 0.37 |
| nano | 0.0374 | 25.64 | 8.76 | 0.64 | 0.0154 | 64.85 | 39.86 | 0.66 | |
| micro | 0.1029 | 32.20 | 10.34 | 0.95 | 0.0117 | 63.08 | 35.96 | 0.43 | |
| SVM-Type2 | pico | 0.0652 | 35.79 | 19.13 | 0.80 | 0.0652 | 26.99 | 0.29 | 0.80 |
| nano | 0.0370 | 28.09 | 8.06 | 0.58 | 0.0379 | 45.05 | 22.28 | 0.68 | |
| micro | 0.1771 | 35.24 | 11.77 | 0.90 | 0.0485 | 50.14 | 21.60 | 0.62 | |
| SVM-Type3 | pico | 0.0655 | 14.88 | 7.19 | 0.80 | 0.0794 | 39.83 | 13.89 | 0.68 |
| nano | 0.0387 | 31.44 | 9.42 | 0.56 | 0.0385 | 57.71 | 34.14 | 0.59 | |
| micro | 0.1166 | 38.53 | 13.29 | 0.94 | 0.0800 | 58.09 | 23.89 | 0.87 | |
| Models | Size Class | RMS | APD% | RPD% | r2 |
|---|---|---|---|---|---|
| Three-component model | micro | 0.2462 | 295.6 | 288.3 | 0.66 |
| nano | 0.0630 | 262.5 | 204.8 | 0.28 | |
| pico | 0.2281 | 54.94 | 5.051 | 0.53 | |
| SVM-Bricaud95 | micro | 0.0981 | 105.4 | 56.7 | 0.66 |
| nano | 0.0740 | 181.4 | 96 | 0.35 | |
| pico | 0.2516 | 56.28 | 18.81 | 0.57 | |
| SVM-SCM | micro | 0.1077 | 364.6 | 326.7 | 0.68 |
| nano | 0.0558 | 262.2 | 195.3 | 0.11 | |
| pico | 0.3372 | 51.97 | 38.99 | 0.39 |
| This paper | 0.319 | 3.018 | 0.255 | 2.466 |
| Brewin et al. (2010) | 0.775 | 1.152 | 0.146 | 5.118 |
| Variable | Bricaud95 | SCM | ||||
|---|---|---|---|---|---|---|
| RMS | APD% | MR | RMS | APD% | MR | |
| aph(λ)/aph(412) | 0.0685 | 6.70 | 0.99 | 0.2402 | 23.70 | 0.79 |
| aph(λ)/aph(490) | 0.1324 | 23.20 | 1.23 | 0.4218 | 71.25 | 1.66 |
| aph(λ)/aph(510) | 0.1340 | 47.41 | 1.46 | 0.5146 | 159.51 | 2.47 |
| aph(λ)/aph(555) | 0.0726 | 117.81 | 1.77 | 0.5863 | 609.62 | 7.36 |
| aph(λ)/aph(670) | 0.1793 | 64.72 | 1.55 | 0.5142 | 181.73 | 2.64 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deng, L.; Zhou, W.; Cao, W.; Zheng, W.; Wang, G.; Xu, Z.; Li, C.; Yang, Y.; Hu, S.; Zhao, W. Retrieving Phytoplankton Size Class from the Absorption Coefficient and Chlorophyll A Concentration Based on Support Vector Machine. Remote Sens. 2019, 11, 1054. https://doi.org/10.3390/rs11091054
Deng L, Zhou W, Cao W, Zheng W, Wang G, Xu Z, Li C, Yang Y, Hu S, Zhao W. Retrieving Phytoplankton Size Class from the Absorption Coefficient and Chlorophyll A Concentration Based on Support Vector Machine. Remote Sensing. 2019; 11(9):1054. https://doi.org/10.3390/rs11091054
Chicago/Turabian StyleDeng, Lin, Wen Zhou, Wenxi Cao, Wendi Zheng, Guifen Wang, Zhantang Xu, Cai Li, Yuezhong Yang, Shuibo Hu, and Wenjing Zhao. 2019. "Retrieving Phytoplankton Size Class from the Absorption Coefficient and Chlorophyll A Concentration Based on Support Vector Machine" Remote Sensing 11, no. 9: 1054. https://doi.org/10.3390/rs11091054
APA StyleDeng, L., Zhou, W., Cao, W., Zheng, W., Wang, G., Xu, Z., Li, C., Yang, Y., Hu, S., & Zhao, W. (2019). Retrieving Phytoplankton Size Class from the Absorption Coefficient and Chlorophyll A Concentration Based on Support Vector Machine. Remote Sensing, 11(9), 1054. https://doi.org/10.3390/rs11091054