2.1. System
Figure 1 details the proposed approach. Our UAV is a quadrotor called the AscTec Pelican (manufactured by Intel’s Ascending Technologies (
https://robots.ros.org/astec-pelican-and-hummingbird/)). This UAV comes with a C++ software development kit (SDK) that enables onboard code development with ease. A high-level Atom Intel processor (HLP) offers enough computing power to run solutions in real-time, whereas a low-level processor (LLP) handles the sensor data fusion and rotor control with an update rate of 1kHz. As shown in
Figure 1, we developed a closed-loop attitude controller to drive the low-level rotor’s control running in the LLP. This control method depends on the dynamics of the UAV to properly reject wind disturbances and keep the UAV steady. During flight, a dataset of multispectral imagery is precisely collected aiming at the above-ground estimation of nitrogen by using machine learning methods. In previous work [
35], our UAV system was tested during two years of in-field experiments with the aim of estimating above ground biomass accumulation. Here, we have extended the capabilities of the system in
Figure 1 by developing and integrating novel methods for UAV flight control, multispectral imagery segmentation for VI feature extraction, and their impact on nitrogen estimation using machine learning algorithms.
The UAV was equipped with the Parrot Sequoia multispectral sensor (
https://www.parrot.com/business-solutions-us/parrot-professional/parrot-sequoia) fabricated with 4 separate bands: green, red, red-edge, and near-infrared. The camera offers a resolution of
for each independent spectral sensor, yielding a crop-to-image resolution of 1.33 cm/pixel according to the flying altitude of 20 m. In addition, the multispectral camera comes with a radiometric calibration target that enables offline reflectance calibration across the spectrum of light captured by the camera, and an onboard irradiance sensor designed to correct images for illumination differences, all of which enables its outstanding performance in cloudy conditions, as evaluated in [
36]. In our application, this calibration procedure was executed prior to any flight mission of the UAV. Additional sensors such as an IMU, magnetometer, and GPS are also embedded within the sunshine sensor.
Figure 2a depicts the mounted camera setup, while
Table 1 details the general specifications of our system.
2.2. Rice Crops
The crops were designed with 3 spatial repetitions containing 8 contrasting rice genotypes in terms of N concentration, biomass accumulation, and flowering: FED50, MG2, ELWEE, NORUNKAN, IR64, AZUCENA, UPLRI7, and LINE 23. These rice varieties have a phenological cycle ranging between 86–101 days , as detailed in
Figure 3b. The experimental design consisted of six months of in-field sampling with three different planting crops. For each experiment, the amount of applied nitrogen and water was modified as follows:
Experiment 1: 200 kgha of nitrogen and limited crop irrigation.
Experiment 2: 200 kgha of nitrogen with permanent crop irrigation.
Experiment 3: 100 kgha of nitrogen with permanent crop irrigation.
Experimental trials were conducted during the dry season. To assess the effects of limited and permanent irrigation on the crops, leaf temperature (MultispeQ (
https://www.photosynq.com)), and soil humidity (AquaPro (
http://aquaprosensors.com)) were constantly monitored. Fertilizers were applied accordingly in order to maintain the crops through the phenological cycle. Given that, the same amount of fertilizers were applied for the experiments: 60 kgha
of
(diammonium phosphate) and 130 kgha
of
(potassium chloride), as detailed in the experimental protocol available in the
Supplementary Materials section.
Figure 2 details some characteristics of the crop. As shown in plots (a), (b), the length of a plot was about 2.75 m with a distance between plants of 25 cm and 30 cm between rows. Within each plot, we defined 6 sampled areas with 1 linear meter in length (containing four plants). A ground-truth was defined based on the direct measurements of plant chlorophyll using a SPAD 502 Plus meter (Konica-Minolta) over these sampled areas. Measurements from the crop were obtained during three stages of rice growth: vegetative, reproductive, and ripening.
Figure 3b details the specifications of the crop experiments reported here.
Table 2 details how the dataset was collected, in which the SPAD value corresponds to the average of 24 measurements conducted over each plot, as depicted in
Figure 3b. All ground-truth data are available through the Open Science Framework in the
Supplementary Materials section. On the other hand,
Figure 3a shows the linear correlation used to relate the measured SPAD value with the leaf-blade N concentration [
37].
As mentioned, vegetation indices are widely used to quantify both plant and soil variables by associating certain spectral reflectances that are highly related to variations in canopy chemical components such as nitrogen. From the extensive list of vegetation indices (VIs) available [
38,
39,
40,
41], we selected 7 VIs with sufficient experimental evidence and quantitative trait loci (QTL)-based characterization regarding their high correlation with rice nitrogen [
42].
Table 3 details the selected VIs. These formulas are applied in different wavelengths for taking into account the changes in canopy color, since several factors affect the spectral reflectances of the crop: solar radiation, plant morphology and color, leaf angles, undergrowth, soil characteristics, and water.
In our system, the parrot sequoia camera has a solar radiation sensor that compensates the light variations in the canopy. The change in the rice canopy color is perhaps the most notable variation during the phenological cycle. In the vegetative stage, the green color is predominant whereas in the reproductive stage, panicle formation starts and thus yellow features appear in the images. In ripening, the maturation of the plants occur while the leaves begin to senesce, turning the yellow color predominant. These changes can be observed in
Figure 2c. Furthermore, different wavelengths of light have a different level of plant absorption depending on the leaf composition given by several genetic traits. In particular, the relation between the selected VIs in
Table 3 with the photosynthetic activity and canopy structural properties has allowed the association of certain spectral reflectances that are highly related to the physico-chemical canopy N variations in plants, especially the green, red, red-edge, and near infrared bands.
The selected VIs exhibit a strong dependence on the NIR reflectance due to leaf chlorophyll absorption, providing an accurate approach to determine the health status of the plants and the canopy N. Most of the existing body of research focused on multispectral-based N estimations [
14,
21,
40,
42,
43], combine the information provided by several vegetation indices in order to avoid saturation issues. For instance, the NDVI, which is one of the most common VIs used, tends to saturate with dense vegetation. In turn, the NDVI alone is not accurate during the reproductive and ripening stages of rice growth. Here, we combine several VIs across the crop stages, to ensure data on wavelengths located in the red-edge and another spectral reflectances that accurately express the healthy status of the leaves (higher NIR and green-band readings).
2.3. UAV Stabilization Control
The estimation of the canopy N requires the precise extraction of Vegetative Index features from the acquired multispectral imagery. Capturing aerial images during UAV hovering that accurately matches with the GPS positions of the SPAD measurements at ground-level is essential.
As previously detailed in
Figure 1, three closed-loop controllers are needed to regulate: (i) the
X-
Y position based on GPS feedback, (ii) the
Z altitude based on barometric pressure and laser readings (pointing downwards), and (iii) the
attitude based on IMU data. The UAV is constantly subjected to wind disturbances that cause unsteady angular motions and therefore imprecise trajectory tracking. Consequently, aerial imagery captured across the crop with the UAV system is affected. To overcome this issue, we replaced both roll and pitch PID-based controllers by a robust nonlinear backstepping (BS) control.
The classical BS method has several advantages. It explicitly takes into account the nonlinearities of the UAV model defined in Equation (
A2) (see
Appendix A), and most importantly, allows the incorporation of a virtual control law to regulate angular accelerations. For this, we have derived a desired acceleration function (DAF) for roll and pitch. This enhanced controller is called backstepping+DAF (BS+DAF). Our goal is to use the dynamics equations of motion (EoM) defined in Algorithm A1 within the control law in order to compensate for abrupt angular acceleration changes, concretely in roll and pitch. The DAF terms improve the responsiveness of the control law to external perturbative forces.
Appendix B details the control law derivation for roll and pitch.
The BS + DAF control supports on the Lyapunov stability concept that guarantee asymptotic stabilization around the equilibrium points. For our application, we require both roll a pitch angles to remain in zero while the UAV is hovering above the crop for capturing multispectral imagery, i.e., and . Otherwise, the set-point references for both roll and pitch controllers are defined by the X-Y position controller.
As previously mentioned, high wind-speed perturbations affect the UAV during hovering. Nonetheless, our controller law is sensitive to angular acceleration changes caused by external wind disturbances, thanks to both error dynamics (
) and the DAF terms (
) in Equation (
A18) (
Appendix B). Given that, we present how to model the effects of the wind over the UAV aerodynamics, by following the wind effect model (WEM) developed by [
44].
Figure 4 details the control architecture and how the WEM module has been incorporated as an external disturbance acting on the UAV body frame.
The WEM defines the relationship between the oblique airflow acting on a propeller and the resulting forces and moments applied onto the UAV. In our quadrotor system, the oblique airflow form and angle of 90
o with the propeller azimuth angle. In terms of aerodynamics, the incoming airflow induces an increase in vertical thrust (
), a side force (F
), and a pitching moment (M
). The WEM presented by [
44] uses blade element momentum theory to determine the aforementioned effects. Here, we are interested in incorporating the effects caused to the thrust and the pitching moment (M
) as a function of the magnitude of a given wind speed vector
and the rotational speed
of each propeller driven by the rotor’s model. In this regard, the WEM is defined as:
The angle
determines the direction of the
vector. Depending on the direction of the applied relative wind, certain propellers will be more affected by this external disturbance. Tran et al., in [
44,
45], propose the use of a weight function that assigns values between 0 to 1, where the maximum value of 1 means the propeller is directly exposed to the wind. In Equation (
1),
C corresponds to the weighting vector that affects the thrust
T generated by each propeller. The parameter
l is the distance between non-adjacent propellers and
is:
Therefore, the scale of the weighting vector
C is defined based on the magnitude of the applied wind speed vector and the corresponding direction (the
angle between the relative wind vector pointing towards the UAV body frame). In addition, the rotor’s speeds
are required to calculate the augmented thrusts
T and torques
(weighted by
C). Further details on this model can be found in [
44,
45].
Section 3.1 and
Section 3.3 present the results regarding the impact of precise UAV positioning on the accurate estimation of the canopy N through the entire phenological cycle.
2.5. Machine Learning for N Estimations
As detailed in
Table 2, the ground-truth for training machine learning (ML) algorithms was defined based on the direct measurements of plant chlorophyll using a SPAD 502 Plus meter (Konica-Minolta) over these sampled areas, as depicted in
Figure 1. Datasets contain the measured SPAD value that directly correlates with the leaf-blade N concentrations by following the linear correlation [
37] defined in
Figure 3a. In this regard, measurements from the crop were obtained during three stages of rice growth: vegetative, reproductive, and ripening, in which 3 trials were conducted per crop stage. These datasets are the result of 10 flights per trial, capturing around 500 images, and yielding a database of
images per stage. Since 3 trials were conducted per crop stage, around
images were processed in this work.
Figure 3b details the experimental specifications.
The ML methods were trained with a set of images accounting for the
of the entire database. For the final estimations of leaf nitrogen, we used the remaining
of the database (testing phase of the ML methods). The entire imagery dataset and the ground-truth available in the
Supplementary Materials section. Here, we report on the use of classical ML methods based on multi-variable linear regressions (MLR), support vector machines (SVM), and artificial neural networks (NN) for the estimation of the canopy N.
For MLR models, the VIs from
Table 3 were combined by following the formula:
, where the parameter
changes accordingly to the growth stage of the plant, by weighting each VI independently. In this regard, each crop stage will have an independent MLR model that linearly fits the N content.
With the aim of improving the adaptability of the estimation models by considering several nonlinear effects (dense vegetation, optical properties of the soil, and changes in canopy color), this section presents the estimation results using support vector machines (SVM) and artificial neural networks (NN). SVM models were used with different kernel functions with the aim of determining the proper mathematical function for mapping the input data. Six different kernels based on linear, quadratic, cubic, and Gaussian models will be tested for each crop stage independently.
Contrarily to the MLR and SVM methods, in which N estimation models are determined for each crop stage independently, neural networks enable the combination of the entire dataset (SPAD values and VI extracted features) into a single model that fits for the entire crop phenological stages. Several non-linear training functions are tested with different hidden layers. In addition, we discarded the use of deep-learning methods such as convolutional neural networks (CNN), due to the high computational costs associated to the pooling through lots of hidden layers in order to detect data features. For this application, we use well-known vegetative index features (reported in
Table 3) that have been widely used and validated in the specialized literature [
14,
40,
42]. Other image-based features such as color, structure, and morphology do not work well with multispectral imagery of 1.2 mega-pixel in resolution, compared to the 16 mega-pixel in the RGB image. In fact, the main advantage of using VIs (as features for training), relies on having information at different wavelengths, providing key information of the plant health status related to N.
In
Section 3.3, we report a comprehensive comparison among multi-variable linear regressions (MLR), support vector machines (SVM), and artificial neural networks (NN) for the estimation of the canopy N. We used three metrics based on the root mean square error (
), Pearson’s linear correlation (
r), and coefficient of determination (
), detailed as follows:
where
n is the total number of samples,
N is the measured value (SPAD scale cf.
Figure 3a),
is the estimated nitrogen, and
is the mean of the
N values. In this application, the Pearson’s metric indicates the linear relationship between the estimated value of N VS the measured one (ground-truth). On the other hand, the
metric is useful since it penalizes the variance of the estimated value from the measured one.
As mentioned, the ML models require the calculation of the VI formulas presented in
Table 3, in order to determine the input feature vector. Every image captured by the UAV is geo-referenced with the DGPS system with a position accuracy of 35 [cm] (see
Table 1). Given that, aerial imagery is registered by matching consecutive frames according to a homography computed with the oriented features from accelerated segment test (FAST) and rotated binary robust independent elementary feature (BRIEF) computer vision techniques [
47]. The UAV path is planned by ensuring an image overlapping of
, which allows for the precise matching with the GPS coordinates of the ground-level markers to ensure a proper comparison between the aerial-estimations and ground-measurements of plant N. Part of this procedure is described in previous work reported in [
48]. In turn, the metrics described in Equation (
10) report on the performance of the ML models based on the aerial-ground data matching of canopy N. The aforementioned geo-referencing process is conducted by using the affine transformation, a 1st order polynomial function that relates the GPS latitude and longitude values with the pixel coordinates within the image. This procedure is also detailed in previous work reported in [
48].
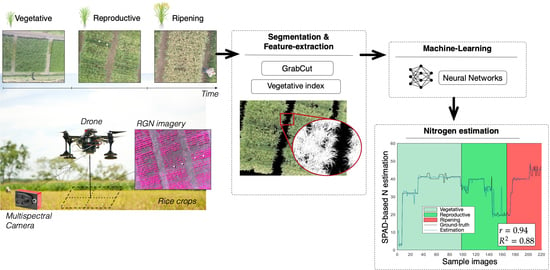
Figure 5 details the procedure required by the machine-learning models. After registration, images are segmented by using the proposed GrabCut method. Although all pixels in the image are evaluated in Algorithm 2, we use pixel clusters of
to calculate the VI formulas, as shown in
Figure 5a. After segmentation, pixels representing the rice canopy are separated from the background, as shown in
Figure 5b. All vegetation indices are calculated as the average within each image sub-region.
At canopy-level, several factors affect the spectral reflectances of the crop: solar radiation and weather conditions, plant morphology and color, leaf angles, undergrowth, soil characteristics, and ground water layers. As mentioned in
Section 2.1, the multispectral camera comes with an integrated sunshine sensor to compensate light variations in the resultant image. In addition, the GrabCut segmentation method deals with the filtering of undergrowth and other soil noises. Despite that, it remains crucial to validate the accuracy of the selected VIs, since the estimation of N depends on the accuracy and reliability of the extracted features. In this regard,
Figure 5c presents several VI calculations in order to analyze the variance of the VI features through the entire phenological cycle of the crop. For this test, we calculated the VI formulas from 360
random images per crop stage under different environmental conditions. We show the most representative VIs to nitrogen variations: simple ratio (SR), normalized difference vegetation index (NDVI), green normalized difference vegetation index (GNDVI), and soil-adjusted vegetation index (MSAVI). As observed, the low variance exhibited through the entire phenological cycle allows us to use the VI features as inputs to the ML models detailed in
Figure 5d.