UAV Laser Scans Allow Detection of Morphological Changes in Tree Canopy
Abstract
:1. Introduction
2. Materials and Methods
2.1. Research Area
2.2. Data Acquisition
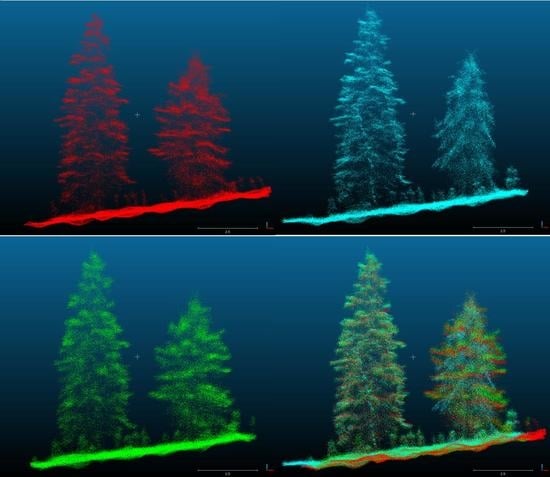
2.3. Data Processing
2.4. Field Data Collection
2.5. Evaluation
3. Results
3.1. The Detected Trees
3.2. Automatic Identification of Trees with Habitus Change
4. Discussion
- In cases where portions of the roots are still alive, the existing root connection to neighboring living trees can mediate responses to environmental conditions, even in the case of fatal loss of assimilation apparatus. Fungal interconnections support not only the transfer of water [44] but also carbon and nitrogen compounds [45]. Moreover, mycorrhizal connections act as conduits for signaling between plants [46]. It is highly probable that these conduits can also transport signals associated with solar radiation. Water uptake is driven by the transpiration demands of the assimilation apparatus. In the case of dead trees, water uptake could be due to the transpiration apparatus of the living tree and/or water efflux through the lenticels in the periderm [47,48]. This hypothesis is supported by the close proximity of living blue spruce to all dead trees in the study.
- Water uptake from the soil by any surviving roots of the dead trees without any support from other trees or fungus. This hypothesis supports the highest amount of precipitation during the winter period, e.g., with a surplus of water during the early spring and autumn. The water uptake through root systems which have been killed was described by [49].
- The branch shifting during the year could be related to the branch length [50]. This observation using an RTM approach in a mixed broadleaf forest confirmed the sagging of a branch and on the opposite branch stub rising after losing its terminal part, simply as the total branch weight decreased.
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Surovỳ, P.; Kuželka, K. Acquisition of forest attributes for decision support at the forest enterprise level using remote-sensing techniques—A review. Forests 2019, 10, 273. [Google Scholar] [CrossRef] [Green Version]
- Næsset, E.; Gobakken, T. Estimating forest growth using canopy metrics derived from airborne laser scanner data. Remote Sens. Environ. 2005, 96, 453–465. [Google Scholar] [CrossRef]
- Means, J.E.; Acker, S.A.; Fitt, B.J.; Renslow, M.; Emerson, L.; Hendrix, C.J. Predicting forest stand characteristics with airborne scanning lidar. Photogramm. Eng. Remote Sens. 2000, 66, 1367–1371. [Google Scholar]
- Packalen, P.; Strunk, J.; Packalen, T.; Maltamo, M.; Mehtätalo, L. Resolution dependence in an area-based approach to forest inventory with airborne laser scanning. Remote Sens. Environ. 2019, 224, 192–201. [Google Scholar] [CrossRef]
- Verma, N.K.; Lamb, D.W.; Reid, N.; Wilson, B. An allometric model for estimating DBH of isolated and clustered Eucalyptus trees from measurements of crown projection area. For. Ecol. Manag. 2014, 326, 125–132. [Google Scholar] [CrossRef]
- Darwin, C. The Power of Movement in Plants; Appleton: New York, NY, USA, 1897. [Google Scholar]
- McClung, C.R. The plant circadian oscillator. Biology 2019, 8, 14. [Google Scholar] [CrossRef] [Green Version]
- Zobel, B.J.; Jett, J.B. Genetics of Wood Production; Springer Science & Business Media: Cham, Switzerland, 2012; ISBN 3642795145. [Google Scholar]
- Pokorny, R.; Tomaskova, I.; Marek, M.V. The effects of elevated atmospheric [CO2] on Norway spruce needle parameters. Acta Physiol. Plant. 2011, 33, 2269–2277. [Google Scholar] [CrossRef]
- Puttonen, E.; Briese, C.; Mandlburger, G.; Wieser, M.; Pfennigbauer, M.; Zlinszky, A.; Pfeifer, N. Quantification of overnight movement of birch (Betula pendula) branches and foliage with short interval terrestrial laser scanning. Front. Plant Sci. 2016, 7, 222. [Google Scholar] [CrossRef] [Green Version]
- Zlinszky, A.; Molnár, B.; Barfod, A.S. Not all trees sleep the same—High temporal resolution terrestrial laser scanning shows differences in nocturnal plant movement. Front. Plant Sci. 2017, 8, 8. [Google Scholar] [CrossRef] [Green Version]
- Brede, B.; Lau, A.; Bartholomeus, H.M.; Kooistra, L. Comparing RIEGL RiCOPTER UAV LiDAR derived canopy height and DBH with terrestrial LiDAR. Sensors 2017, 17, 2371. [Google Scholar] [CrossRef]
- Wieser, M.; Mandlburger, G.; Hollaus, M.; Otepka, J.; Glira, P.; Pfeifer, N. A case study of UAS borne laser scanning for measurement of tree stem diameter. Remote Sens. 2017, 9, 1154. [Google Scholar] [CrossRef] [Green Version]
- Liu, K.; Shen, X.; Cao, L.; Wang, G.; Cao, F. Estimating forest structural attributes using UAV-LiDAR data in Ginkgo plantations. ISPRS J. Photogramm. Remote Sens. 2018, 146, 465–482. [Google Scholar] [CrossRef]
- Peng, X.; Li, X.; Wang, C.; Zhu, J.; Liang, L.; Fu, H.; Du, Y.; Yang, Z.; Xie, Q. SPICE-based SAR tomography over forest areas using a small number of P-band airborne F-SAR images characterized by non-uniformly distributed baselines. Remote Sens. 2019, 11, 975. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Lehtomäki, M.; Liang, X.; Pyörälä, J.; Kukko, A.; Jaakkola, A.; Liu, J.; Feng, Z.; Chen, R.; Hyyppä, J. Is field-measured tree height as reliable as believed—A comparison study of tree height estimates from field measurement, airborne laser scanning and terrestrial laser scanning in a boreal forest. ISPRS J. Photogramm. Remote Sens. 2019, 147, 132–145. [Google Scholar] [CrossRef]
- Jurjević, L.; Liang, X.; Gašparović, M.; Balenović, I. Is field-measured tree height as reliable as believed—Part II, A comparison study of tree height estimates from conventional field measurement and low-cost close-range remote sensing in a deciduous forest. ISPRS J. Photogramm. Remote Sens. 2020, 169, 227–241. [Google Scholar] [CrossRef]
- Lefsky, M.; Harding, D.; Blair, J.; Parker, G. Laser altimeter canopy height profiles: Methods and validation for closed-canopy, broadleaf forests. Remote Sens. Environ. 2001, 76, 283–297. [Google Scholar]
- Næsset, E.; Gobakken, T. Estimation of above- and below-ground biomass across regions of the boreal forest zone using airborne laser. Remote Sens. Environ. 2008, 112, 3079–3090. [Google Scholar] [CrossRef]
- Solodukhin, V.I.; Zhukov, A.Y.; Mazhugin, I.N.; Narkevich, V.I. Metody Izuchenija Vertikal’nyh Sechenij Drevostoev (Method of Study of Vertical Sections of Forest Stands); Leningrad Scientific Research Institute of Forestry: Leningrad, Russia, 1976; 55p. [Google Scholar]
- Nelson, R.; Krabill, W.; MacLean, G. Determining forest canopy characteristics using airborne laser data. Remote Sens. Environ. 1984, 15, 201–212. [Google Scholar] [CrossRef]
- Rosati, A.; Paoletti, A.; Caporali, S.; Perri, E. The role of tree architecture in super high density olive orchards. Sci. Hortic. 2013, 161, 24–29. [Google Scholar] [CrossRef]
- Lau, A.; Bentley, L.P.; Martius, C.; Shenkin, A.; Bartholomeus, H.; Raumonen, P.; Malhi, Y.; Jackson, T.; Herold, M. Quantifying branch architecture of tropical trees using terrestrial LiDAR and 3D modelling. Trees Struct. Funct. 2018, 32, 1219–1231. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez de Tanago, J.; Lau, A.; Bartholomeus, H.; Herold, M.; Avitabile, V.; Raumonen, P.; Martius, C.; Goodman, R.C.; Disney, M.; Manuri, S.; et al. Estimation of above-ground biomass of large tropical trees with terrestrial LiDAR. Methods Ecol. Evol. 2018, 9, 223–234. [Google Scholar] [CrossRef] [Green Version]
- Jaakkola, A.; Hyyppä, J.; Kukko, A.; Yu, X.; Kaartinen, H.; Lehtomäki, M.; Lin, Y. A low-cost multi-sensoral mobile mapping system and its feasibility for tree measurements. ISPRS J. Photogramm. Remote Sens. 2010, 65, 514–522. [Google Scholar] [CrossRef]
- Wallace, L.; Lucieer, A.; Watson, C.; Turner, D. Development of a UAV-LiDAR system with application to forest inventory. Remote Sens. 2012, 4, 1519–1543. [Google Scholar] [CrossRef] [Green Version]
- Panagiotidis, D.; Abdollahnejad, A.; Surový, P.; Chiteculo, V. Determining tree height and crown diameter from high-resolution UAV imagery. Int. J. Remote Sens. 2017, 38, 2392–2410. [Google Scholar] [CrossRef]
- Lowe, D.G. Method and Apparatus for Identifying Scale Invariant Features in an Image. U.S. Patent US6711293B1, 23 March 2004. [Google Scholar]
- Lee, S.K.; Fatoyinbo, T.; Qi, W.; Hancock, S.; Armston, J.; Dubayah, R. GEDI and TanDEM-X fusion for 3D forest structure parameter retrieval. Int. Geosci. Remote Sens. Symp. 2018, 2018, 380–382. [Google Scholar]
- Qi, W.; Dubayah, R.O. Combining Tandem-X InSAR and simulated GEDI lidar observations for forest structure mapping. Remote Sens. Environ. 2016, 187, 253–266. [Google Scholar] [CrossRef]
- Mitchard, E.T.A. The tropical forest carbon cycle and climate change. Nature 2018, 559, 527–534. [Google Scholar] [CrossRef]
- Baccini, A.; Walker, W.; Carvalho, L.; Farina, M.; Houghton, R.A. Response to Comment on “Tropical forests are a net carbon source based on aboveground measurements of gain and loss”. Science 2019, 363, 230–234. [Google Scholar] [CrossRef] [Green Version]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Šrámek, V.; Slodičák, M.; Lomský, B.; Balcar, V.; Kulhavý, J.; Hadaš, P.; Pulkráb, K.; Šišák, L.; Pěnička, L.; Sloup, M. The Ore Mountains: Will Successive Recovery of Forests from Lethal Disease Be Successful. Mt. Res. Dev. 2008, 28, 216–221. [Google Scholar] [CrossRef] [Green Version]
- Černý, K.; Pešková, V.; Soukup, F.; Havrdová, L.; Strnadová, V.; Zahradník, D.; Hrabětová, M. Gemmamyces bud blight of Picea pungens: A sudden disease outbreak in Central Europe. Plant Pathol. 2016, 65, 1267–1278. [Google Scholar] [CrossRef] [Green Version]
- Riegl. RIEGL VUX-SYS. 2019, Volume 4. Available online: http://www.riegl.com/uploads/tx_pxpriegldownloads/RIEGL_VUX-SYS_Datasheet_2020-10-02_01.pdf (accessed on 20 November 2020).
- Czech Republic. Letecký Předpis L 2 Pravidla Létání. 2014. Available online: https://aim.rlp.cz/predpisy/predpisy/dokumenty/L/L-2/data/print/L-2_cely.pdf (accessed on 20 November 2020).
- Westoby, M.J.; Brasington, J.; Glasser, N.F.; Hambrey, M.J.; Reynolds, J.M. “Structure-from-Motion” photogrammetry: A low-cost, effective tool for geoscience applications. Geomorphology 2012, 179, 300–314. [Google Scholar] [CrossRef] [Green Version]
- Lisein, J.; Pierrot-Deseilligny, M.; Bonnet, S.; Lejeune, P. A photogrammetric workflow for the creation of a forest canopy height model from small unmanned aerial system imagery. Forests 2013, 4, 922–944. [Google Scholar] [CrossRef] [Green Version]
- Bosshard, W.; Müller, E. Kronenbilder: Mit Nadel-und Blattverlustprozenten; Eidgenössische Anstalt für das Forstliche Versuchswesen: Birmensdorf, Switzerland, 1986. [Google Scholar]
- Pekár, S.; Brabec, M. Modern Analysis of Biological Data: Generalized Linear Models in R; Masarykova Univerzita: Brno, Czech Republic, 2016; ISBN 8021081066. [Google Scholar]
- Hyyppä, E.; Hyyppä, J.; Hakala, T.; Kukko, A.; Wulder, M.A.; White, J.C.; Pyörälä, J.; Yu, X.; Wang, Y.; Virtanen, J.P.; et al. Under-canopy UAV laser scanning for accurate forest field measurements. ISPRS J. Photogramm. Remote Sens. 2020, 164, 41–60. [Google Scholar] [CrossRef]
- Hyyppä, E.; Yu, X.; Kaartinen, H.; Hakala, T.; Kukko, A.; Vastaranta, M.; Hyyppä, J. Comparison of Backpack, Handheld, Under-Canopy UAV, and Above-Canopy UAV Laser Scanning for Field Reference Data Collection in Boreal Forests. Remote Sens. 2020, 12, 3327. [Google Scholar] [CrossRef]
- Plamboeck, A.H.; Dawson, T.E.; Egerton-Warburton, L.M.; North, M.; Bruns, T.D.; Querejeta, J.I. Water transfer via ectomycorrhizal fungal hyphae to conifer seedlings. Mycorrhiza 2007, 17, 439–447. [Google Scholar] [CrossRef] [PubMed]
- Teste, F.P.; Simard, S.W.; Durall, D.M. Role of mycorrhizal networks and tree proximity in ectomycorrhizal colonization of planted seedlings. Fungal Ecol. 2009, 2, 21–30. [Google Scholar] [CrossRef]
- Babikova, Z.; Gilbert, L.; Bruce, T.J.A.; Birkett, M.; Caulfield, J.C.; Woodcock, C.; Pickett, J.A.; Johnson, D. Underground signals carried through common mycelial networks warn neighbouring plants of aphid attack. Ecol. Lett. 2013, 16, 835–843. [Google Scholar] [CrossRef]
- Baret, M.; DesRochers, A. Root connections can trigger physiological responses to defoliation in nondefoliated aspen suckers. Botany 2011, 89, 753–761. [Google Scholar] [CrossRef]
- Simard, S.W.; Jones, M.D.; Durall, D.M. Carbon and nutrient fluxes within and between mycorrhizal plants. In Mycorrhizal Ecology; Springer: Cham, Switzerland, 2003; pp. 33–74. [Google Scholar]
- Kramer, P.J. The intake of water through dead root systems and its relation to the problem of absorption by transpiring plants. Am. J. Bot. 1933, 20, 481–492. [Google Scholar] [CrossRef]
- Putman, E.B.; Popescu, S.C.; Eriksson, M.; Zhou, T.; Klockow, P.; Vogel, J.; Moore, G.W. Detecting and quantifying standing dead tree structural loss with reconstructed tree models using voxelized terrestrial lidar data. Remote Sens. Environ. 2018, 209, 52–65. [Google Scholar] [CrossRef]
- Barak, S.; Tobin, E.M.; Green, R.M.; Andronis, C.; Sugano, S. All in good time: The Arabidopsis circadian clock. Trends Plant Sci. 2000, 5, 517–522. [Google Scholar] [CrossRef]
- Ibañez, C.; Ramos, A.; Acebo, P.; Contreras, A.; Casado, R.; Allona, I.; Aragoncillo, C. Overall Alteration of Circadian Clock Gene Expression in the Chestnut Cold Response. PLoS ONE 2008, 3, e3567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Solomon, O.L.; Berger, D.K.; Myburg, A.A. Diurnal and circadian patterns of gene expression in the developing xylem of Eucalyptus trees. S. Afr. J. Bot. 2010, 76, 425–439. [Google Scholar] [CrossRef] [Green Version]
- Shimano, K. Analysis of the relationship between DBH and crown projection area using a new model. J. For. Res. 1997, 2, 237–242. [Google Scholar] [CrossRef]







| RiCOPTER | Characteristics |
|---|---|
| Total weight | 25 kg |
| Sensors (VUX-1UAV + cameras) | 3.5 kg |
| Batteries (Li-Pol 29.4V 12,500 mAh) | 7.5 kg |
| Maximum flight time | 28 min |
| Maximum horizontal speed | 14 m·s−1 |
| Maximum ascending speed | 5 m·s−1 |
| Maximum descending speed | 2 m·s−1 |
| Estimate | SE | tStat | p-Value | |
|---|---|---|---|---|
| Intercept | 32.884 | 22.659 | 1.451 | 0.1467 |
| A1 | 43.728 | 30.725 | 1.423 | 0.1547 |
| A2 | 6.350 | 4.130 | 1.538 | 0.1241 |
| A5 | −53.701 | 26.340 | −2.039 | 0.0415 |
| A6 | −28.857 | 15.987 | −1.805 | 0.0711 |
| A9 | −15.078 | 8.609 | −1.752 | 0.0798 |
| Dataset | Trees | Change | False Negative | False Positive | Correct |
|---|---|---|---|---|---|
| Training | 204 | 29 | 1 | 2 | 28 |
| Validation | 204 | 14 | 2 | 5 | 12 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Slavík, M.; Kuželka, K.; Modlinger, R.; Tomášková, I.; Surový, P. UAV Laser Scans Allow Detection of Morphological Changes in Tree Canopy. Remote Sens. 2020, 12, 3829. https://doi.org/10.3390/rs12223829
Slavík M, Kuželka K, Modlinger R, Tomášková I, Surový P. UAV Laser Scans Allow Detection of Morphological Changes in Tree Canopy. Remote Sensing. 2020; 12(22):3829. https://doi.org/10.3390/rs12223829
Chicago/Turabian StyleSlavík, Martin, Karel Kuželka, Roman Modlinger, Ivana Tomášková, and Peter Surový. 2020. "UAV Laser Scans Allow Detection of Morphological Changes in Tree Canopy" Remote Sensing 12, no. 22: 3829. https://doi.org/10.3390/rs12223829
APA StyleSlavík, M., Kuželka, K., Modlinger, R., Tomášková, I., & Surový, P. (2020). UAV Laser Scans Allow Detection of Morphological Changes in Tree Canopy. Remote Sensing, 12(22), 3829. https://doi.org/10.3390/rs12223829