Recognition of Banana Fusarium Wilt Based on UAV Remote Sensing
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Field Investigation
2.3. UAV Multi-Spectral Imagery Acquisition
2.4. Data Analysis
2.4.1. Vegetation Indices
2.4.2. Binary Logistic Regression
2.4.3. Accuracy Assessment
3. Results
3.1. Statistical Characteristics of Samples
3.2. Model Fitting with Different Vegetation Indices
3.3. Model Fitting for Different Resolution Imagery
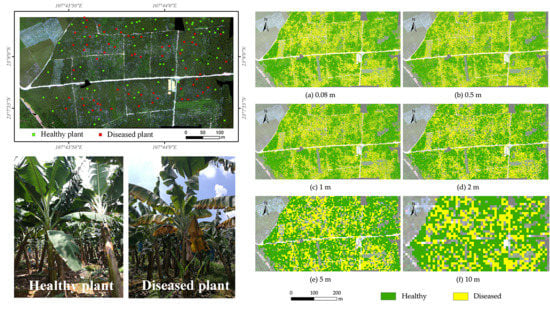
3.4. Mapping Disease Distribution using Imagery with Different Resolutions
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Shen, Z.; Xue, C.; Penton, C.R.; Thomashow, L.S.; Zhang, N.; Wang, B.; Ruan, Y.; Li, R.; Shen, Q. Suppression of banana Panama disease induced by soil microbiome reconstruction through an integrated agricultural strategy. Soil. Biol. Biochem. 2019, 128, 164–174. [Google Scholar] [CrossRef]
- Ordonez, N.; Seidl, M.F.; Waalwijk, C.; Drenth, A.; Kilian, A.; Thomma, B.P.H.J.; Ploetz, R.C.; Kema, G.H.J. Worse comes to worst: Bananas and Panama disease-when plant and pathogen clones meet. PLoS Pathog. 2015, 11, e1005197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van den Berg, N.; Berger, D.K.; Hein, I.; Birch, P.R.; Wingfield, M.J.; Viljoen, A. Tolerance in banana to Fusarium wilt is associated with early up-regulation of cell wall-strengthening genes in the roots. Mol. Plant Pathol. 2007, 8, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Lin, B.; Shen, H. Fusarium oxysporum f. sp. cubense. In Biological Invasions and Its Management in China: Volume 2; Wan, F., Jiang, M., Zhan, A., Eds.; Springer: Singapore, 2017. [Google Scholar]
- Shi, Y.; Huang, W.; Ye, H.; Ruan, C.; Xing, N.; Geng, Y.; Dong, Y.; Peng, D. Partial least square discriminant analysis based on normalized two-stage vegetation indices for mapping damage from rice diseases using PlanetScope datasets. Sensors 2018, 18, 1901. [Google Scholar] [CrossRef] [Green Version]
- Huang, W.; Lamb, D.W.; Niu, Z.; Zhang, Y.; Liu, L.; Wang, J. Identification of yellow rust in wheat using in-situ spectral reflectance measurements and airborne hyperspectral imaging. Precis. Agric. 2007, 8, 187–197. [Google Scholar] [CrossRef]
- Huang, W.; Guan, Q.; Luo, J.; Zhang, J.; Zhao, J.; Liang, D.; Huang, L.; Zhang, D. New optimized spectral indices for identifying and monitoring winter wheat diseases. IEEE J. Sel. Top. Appl. Earth Observ. Remote Sens. 2014, 7, 2516–2524. [Google Scholar] [CrossRef]
- Shi, Y.; Huang, W.; Gonzalez-Moreno, P.; Luke, B.; Dong, Y.; Zheng, Q.; Ma, H.; Liu, L. Wavelet-based rust spectral feature set (WRSFs): A novel spectral feature set based on continuous wavelet transformation for tracking progressive host-pathogen interaction of yellow rust on wheat. Remote Sens. 2018, 10, 525. [Google Scholar] [CrossRef] [Green Version]
- Jin, X.; Jie, L.; Wang, S.; Qi, H.J.; Li, S.W. Classifying wheat hyperspectral pixels of healthy heads and Fusarium head blight disease using a deep neural network in the wild field. Remote Sens. 2018, 10, 395. [Google Scholar] [CrossRef] [Green Version]
- Mahlein, A.K.; Alisaac, E.; Al Masri, A.; Behmann, J.; Dehne, H.W.; Oerke, E.C. Comparison and Combination of Thermal, Fluorescence, and Hyperspectral Imaging for Monitoring Fusarium Head Blight of Wheat on Spikelet Scale. Sensors 2019, 19, 2281. [Google Scholar] [CrossRef] [Green Version]
- Yuan, L.; Pu, R.; Zhang, J.; Wang, J.; Yang, H. Using high spatial resolution satellite imagery for mapping powdery mildew at a regional scale. Precis. Agric. 2016, 17, 332–348. [Google Scholar] [CrossRef]
- Zhao, J.; Xu, C.; Xu, J.; Huang, L.; Zhang, D.; Liang, D. Forecasting the wheat powdery mildew (Blumeria graminis f. Sp tritici) using a remote sensing-based decision-tree classification at a provincial scale. Australas Plant Path. 2018, 47, 53–61. [Google Scholar] [CrossRef]
- Huang, J.; Liao, H.; Zhu, Y.; Sun, J.; Sun, Q.; Liu, X. Hyperspectral detection of rice damaged by rice leaf folder (Cnaphalocrocis medinalis). Comput. Electron. Agric. 2012, 82, 100–107. [Google Scholar] [CrossRef]
- Yang, C.M. Assessment of the severity of bacterial leaf blight in rice using canopy hyperspectral reflectance. Precis. Agric. 2010, 11, 61–81. [Google Scholar] [CrossRef]
- Dhau, I.; Adam, E.; Mutanga, O.; Ayisi, K.; Abdel-Rahman, E.M.; Odindi, J.; Masocha, M. Testing the capability of spectral resolution of the new multispectral sensors on detecting the severity of grey leaf spot disease in maize crop. Geocarto Int. 2018, 33, 1223–1236. [Google Scholar] [CrossRef]
- Zhang, M.; Qin, Z.; Liu, X.; Ustin, S.L. Detection of stress in tomatoes induced by late blight disease in California, USA, using hyperspectral remote sensin. Int. J. Appl. Earth. Obs. Geoinf. 2003, 4, 295–310. [Google Scholar] [CrossRef]
- Jones, C.D.; Jones, J.B.; Lee, W.S. Diagnosis of bacterial spot of tomato using spectral signatures. Comput. Electron. Agric. 2010, 74, 329–335. [Google Scholar] [CrossRef]
- Zheng, Q.; Huang, W.; Cui, X.; Shi, Y.; Liu, L. New spectral index for detecting wheat yellow rust using Sentinel-2 multispectral imagery. Sensors 2018, 18, 868. [Google Scholar] [CrossRef] [Green Version]
- Bravo, C.; Moshou, D.; West, J.; McCartney, A.; Ramon, H. Early disease detection in wheat fields using spectral reflectance. Biosyst. Eng. 2003, 84, 137–145. [Google Scholar] [CrossRef]
- Devadas, R.; Lamb, D.W.; Simpfendorfer, S.; Backhouse, D. Evaluating ten spectral vegetation indices for identifying rust infection in individual wheat leaves. Precis. Agric. 2009, 10, 459–470. [Google Scholar] [CrossRef]
- Oumar, Z.; Mutanga, O. Using WorldView-2 bands and indices to predict bronze bug (Thaumastocoris peregrinus) damage in plantation forests. Int. J. Remote. Sens. 2013, 34, 2236–2249. [Google Scholar] [CrossRef]
- Zhang, J.; Huang, Y.; Yuan, L.; Yang, G.; Chen, L.; Zhao, C. Using satellite multi-spectral imagery for damage mapping of armyworm (Spodoptera frugiperda) in maize at a regional scale. Pest Manag. Sci. 2016, 72, 335–348. [Google Scholar] [CrossRef] [PubMed]
- Dash, J.P.; Watt, M.S.; Pearse, G.D.; Heaphy, M.; Dungey, H.S. Assessing very high resolution UAV imagery for monitoring forest health during a simulated disease outbreak. ISPRS J. Photogramm. Remote Sens. 2017, 131, 1–14. [Google Scholar] [CrossRef]
- Deng, L.; Mao, Z.; Li, X.; Hu, Z.; Yan, Y. UAV-based multispectral remote sensing for precision agriculture: A comparison between different cameras. ISPRS J. Photogramm. Remote Sens. 2018, 146, 124–136. [Google Scholar] [CrossRef]
- Liu, B.; Shi, Y.; Duan, Y.; Wu, W. UAV-based crops classification with joint features from orthoimage and DSM data. Int. Arch. Photogram. Remote Sens. Spat. Inform. Sci. 2018, XLII-3, 1023–1028. [Google Scholar] [CrossRef] [Green Version]
- Liu, K.; Zhou, Q.B.; Wu, W.B.; Xia, T.; Tang, H.J. Estimating the crop leaf area index using hyperspectral remote sensing. J. Integr. Agr. 2016, 15, 475–491. [Google Scholar] [CrossRef] [Green Version]
- Machovina, B.L.; Feeley, K.J.; Machovina, B.J. UAV remote sensing of spatial variation in banana production. Crop Pasture Sci. 2016, 67, 1281–1287. [Google Scholar] [CrossRef]
- Ye, H.; Cui, B.; Huang, S.; Dong, Y.; Huang, W.; Ren, Y.; Guo, A.; Jin, Y. Identification of banana Fusarium wilt disease based on UAV multi-spectral imagery. In Proceedings of the International Conference on Intelligent Agriculture, Beijing, China, 18–21 October 2019. [Google Scholar]
- Li, N.; Xie, G.; Zhou, D.; Zhang, C.; Jiao, C. Remote sensing classification of marsh wetland with different resolution images. J. Resour. Ecol. 2016, 7, 107–114. [Google Scholar]
- Fisher, J.R.; Acosta, E.A.; Dennedy-Frank, P.J.; Kroeger, T.; Boucher, T.M. Impact of satellite imagery spatial resolution on land use classification accuracy and modeled water quality. Remote Sens. Ecol. Conserv. 2018, 4, 137–149. [Google Scholar] [CrossRef]
- Mahlein, A.K.; Oerke, E.C.; Steiner, U.; Dehne, H.W. Recent advances in sensing plant diseases for precision crop protection. Eur. J. Plant Pathol. 2012, 133, 197–209. [Google Scholar] [CrossRef]
- Lee, S.; Pradhan, B. Landslide hazard mapping at Selangor, Malaysia using frequency ratio and logistic regression models. Landslides 2007, 4, 33–41. [Google Scholar] [CrossRef]
- Ozdemir, A. Using a binary logistic regression method and GIS for evaluating and mapping the groundwater spring potential in the Sultan Mountains (Aksehir, Turkey). J. Hydrol. 2011, 405, 123–136. [Google Scholar] [CrossRef]
- IUSS Working Group WRB. World Reference Base for Soil Resources 2006; FAO: Rome, Italy, 2006. [Google Scholar]
- Rouse, J.W.; Haas, R.H.; Schell, J.A.; Deering, D.W. Monitoring vegetation systems in the great plains with ERTS. In Proceedings of the Third ERTS-1 Symposium NASA SP-351, Greenbelt, MD, USA, 10–14 December 1973. [Google Scholar]
- Gitelson, A.; Merzlyak, M.N. Spectral reflectance changes associated with autumn senescence of aesculus–hippocastanum L and acer-platanoides L leaves—spectral features and relation to chlorophyll estimation. J. Plant Physiol. 1994, 143, 286–292. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Gritz, Y.; Merzlyak, M.N. Relationships between leaf chlorophyll content and spectral reflectance and algorithms for non-destructive chlorophyll assessment in higher plant leaves. J. Plant Physiol. 2003, 160, 271–282. [Google Scholar] [CrossRef] [PubMed]
- Gitelson, A.A.; Vina, A.; Ciganda, V.; Rundquist, D.C.; Arkebauer, T.J. Remote estimation of canopy chlorophyll content in crops. Geophys. Res. Lett. 2005, 32, L08403. [Google Scholar] [CrossRef] [Green Version]
- Peñuelas, J.; Inoue, Y. Reflectance indices indicative of changes in water and pigment contents of peanut and wheat leaves. Photosynthetica 1999, 36, 355–360. [Google Scholar] [CrossRef]
- Ramoelo, A.; Skidmore, A.K.; Cho, M.A.; Schlerf, M.; Mathieu, R.; Heitkonig, I.M.A. Regional estimation of savanna grass nitrogen using the red-edge band of the spaceborne RapidEye sensor. Int. J. Appl. Earth. Obs. Geoinf. 2012, 19, 151–162. [Google Scholar] [CrossRef]
- Zhou, X.; Huang, W.; Zhang, J.; Kong, W.; Casa, R.; Huang, Y. A novel combined spectral index for estimating the ratio of carotenoid to chlorophyll content to monitor crop physiological and phenological status. Int. J. Appl. Earth. Obs. Geoinf. 2019, 76, 128–142. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Merzlyak, M.N.; Chivkunova, O.B. Optical properties and nondestructive estimation of anthocyanin content in plant leaves. Photochem. Photobiol. 2001, 74, 38–45. [Google Scholar] [CrossRef]
- Congalton, R.G. A review of assessing the accuracy of classifications of remotely sensed data. Remote Sens. Environ. 1991, 37, 35–46. [Google Scholar] [CrossRef]
- Foody, G.M. Classification accuracy comparison: Hypothesis tests and the use of confidence intervals in evaluations of difference, equivalence and non-inferiority. Remote Sens. Environ. 2009, 113, 1658–1663. [Google Scholar] [CrossRef] [Green Version]
- Tuanmu, M.N.; Viña, A.; Bearer, S.; Xu, W.; Ouyang, Z.; Zhang, H.; Liu, J. Mapping understory vegetation using phenological characteristics derived from remotely sensed data. Remote Sens. Environ. 2010, 114, 1833–1844. [Google Scholar] [CrossRef]
- Dong, X.; Wang, M.; Ling, N.; Shen, Q.; Guo, S. Potential role of photosynthesis-related factors in banana metabolism and defense against Fusarium oxysporum f. sp cubense. Environ. Exp. Bot. 2016, 129, 4–12. [Google Scholar] [CrossRef]
- Clevers, J.G.P.W.; de Jong, S.M.; Epema, G.F.; van der Meer, F.; Bakker, W.H.; Skidmore, A.K.; Addink, E.A. MERIS and the red-edge position. Int. J. Appl. Earth. Obs. Geoinf. 2001, 3, 313–320. [Google Scholar] [CrossRef]
- Dash, J.; Curran, P.J. The MERIS terrestrial chlorophyll index. Int. J. Remote Sens. 2004, 25, 5403–5413. [Google Scholar] [CrossRef]
- Mathew, J.; Jha, V.K.; Rawat, G.S. Application of binary logistic regression analysis and its validation for landslide susceptibility mapping in part of Garhwal Himalaya, India. Int. J. Remote Sens. 2007, 28, 2257–2275. [Google Scholar] [CrossRef]
- Lu, J.; Sun, L.; Huang, W. Research progress in monitoring and forecasting of crop pests and diseases by remote sensing. Remote Sens. Technol. Appl. 2019, 34, 21–32. [Google Scholar]



| Vegetation Index | Description | Formulation | Sensitive Parameter | Reference |
|---|---|---|---|---|
| NDVI | Normalized difference vegetation index | (RNIR–Rred)/(RNIR+Rred) | Leaf area index, green biomass | [35] |
| NDRE | Normalized difference red edge index | (RNIR–RRE)/(RNIR+RRE) | Leaf area index, green biomass | [36] |
| CIgreen | Green chlorophyll index | RNIR/Rgreen–1 | Chlorophyll content | [37] |
| CIRE | Red-edge chlorophyll index | RNIR/RRE–1 | Chlorophyll content | [38] |
| SIPI | Structural independent pigment index | (RNIR–Rblue)/(RNIR–Rred) | Pigment content | [39] |
| SIPIRE | Red-edge structural independent pigment index | (RRE–Rblue)/(RRE–Rred) | Pigment content | [40] |
| CARI | Carotenoid index | RRE/Rgreen–1 | Carotenoid content | [41] |
| ARI | Anthocyanin reflectance index | 1/Rgreen–1/RRE | Anthocyanin content | [42] |
| Experiment | VI | Sample Category | No. of Samples | Mean of VI Value | Std. Deviation | p Value (t-test) |
|---|---|---|---|---|---|---|
| Guangxi site | NDVI | Healthy | 57 | 0.54 | 0.11 | 0.00 |
| Diseased | 63 | 0.34 | 0.14 | |||
| NDRE | Healthy | 57 | 0.20 | 0.08 | 0.00 | |
| Diseased | 63 | 0.02 | 0.09 | |||
| CIgreen | Healthy | 57 | 1.08 | 0.32 | 0.00 | |
| Diseased | 63 | 0.43 | 0.33 | |||
| CIRE | Healthy | 57 | 0.56 | 0.22 | 0.00 | |
| Diseased | 63 | 0.09 | 0.22 | |||
| SIPI | Healthy | 57 | 0.88 | 0.36 | 0.24 | |
| Diseased | 63 | 1.68 | 5.26 | |||
| SIPIRE | Healthy | 57 | 0.58 | 0.71 | 0.25 | |
| Diseased | 63 | 2.07 | 9.77 | |||
| CARI | Healthy | 57 | 0.34 | 0.04 | 0.00 | |
| Diseased | 63 | 0.30 | 0.06 | |||
| ARI | Healthy | 57 | 0.85 | 0.15 | 0.00 | |
| Diseased | 63 | 0.62 | 0.16 | |||
| Hainan site | NDVI | Healthy | 16 | 0.44 | 0.05 | 0.00 |
| Diseased | 19 | 0.36 | 0.06 | |||
| NDRE | Healthy | 16 | 0.35 | 0.10 | 0.00 | |
| Diseased | 19 | 0.12 | 0.09 | |||
| CIgreen | Healthy | 16 | 0.92 | 0.26 | 0.00 | |
| Diseased | 19 | 0.49 | 0.26 | |||
| CIRE | Healthy | 16 | 0.35 | 0.10 | 0.00 | |
| Diseased | 19 | 0.12 | 0.09 | |||
| SIPI | Healthy | 16 | 1.07 | 0.07 | 0.06 | |
| Diseased | 19 | 1.18 | 0.12 | |||
| SIPIRE | Healthy | 16 | 1.11 | 0.11 | 0.04 | |
| Diseased | 19 | 1.23 | 0.16 | |||
| CARI | Healthy | 16 | 0.43 | 0.16 | 0.01 | |
| Diseased | 19 | 0.33 | 0.19 | |||
| ARI | Healthy | 16 | 0.87 | 0.30 | 0.03 | |
| Diseased | 19 | 0.61 | 0.35 |
| VI | Logistic Regression Equation | OA* of the Fitting (%) | Validation Dataset 1 | Validation Dataset 2 | ||
|---|---|---|---|---|---|---|
| OA (%) | Kappa | OA (%) | Kappa | |||
| NDVI | y = –11.851×NDVI+5.373 | 86.3 | 83.3 | 0.66 | 62.9 | 0.22 |
| NDRE | y = –15.775×NDRE+1.802 | 90.5 | 87.5 | 0.75 | 65.7 | 0.39 |
| CIgreen | y = –4.144×CIgreen+3.118 | 89.5 | 87.5 | 0.74 | 74.3 | 0.47 |
| CIRE | y = –6.110×CIRE+1.935 | 91.6 | 91.7 | 0.83 | 80.0 | 0.59 |
| CARI | y = –9.966×CARI+3.172 | 62.1 | 66.7 | 0.35 | 60.0 | 0.21 |
| ARI | y = –7.247×ARI+5.326 | 75.8 | 83.3 | 0.66 | 68.6 | 0.37 |
| Resolution | Logistic Regression Equation | OA* of the Fitting (%) | Validation Dataset 1 | Validation Dataset 2 | ||
|---|---|---|---|---|---|---|
| OA (%) | Kappa | OA (%) | Kappa | |||
| CIRE | ||||||
| 0.5 m | y = –5.826×CIRE+1.987 | 90.5 | 91.7 | 0.83 | 85.7 | 0.71 |
| 1 m | y = –4.896×CIRE+1.645 | 83.2 | 79.2 | 0.60 | 74.3 | 0.48 |
| 2 m | y = –4.178×CIRE+1.475 | 81.1 | 75.0 | 0.53 | 71.4 | 0.41 |
| 5 m | y = –2.854×CIRE+1.027 | 76.8 | 70.8 | 0.42 | 65.7 | 0.30 |
| 10 m | y = –1.817×CIRE+0.761 | 69.5 | 62.5 | 0.25 | 62.9 | 0.24 |
| CIgreen | ||||||
| 0.5 m | y = –3.946×CIgreen+3.166 | 87.4 | 87.5 | 0.75 | 74.3 | 0.48 |
| 1 m | y = –3.266×CIgreen+2.633 | 83.2 | 75.0 | 0.51 | 65.7 | 0.32 |
| 2 m | y = –2.936×CIgreen+2.421 | 78.9 | 75.0 | 0.51 | 62.9 | 0.26 |
| 5 m | y = –1.862×CIgreen+1.552 | 70.5 | 66.7 | 0.35 | 48.6 | 0.01 |
| 10 m | y = –1.158×CIgreen+1.044 | 61.1 | 58.3 | 0.18 | 45.7 | −0.01 |
| Resolution | Healthy Area (ha) | Diseased Area (ha) | Percentage of Diseased Area (%) |
|---|---|---|---|
| CIRE | |||
| 0.08 m | 8.78 | 6.04 | 40.8 |
| 0.5 m | 8.28 | 6.59 | 44.3 |
| 1 m | 8.60 | 6.28 | 42.2 |
| 2 m | 8.38 | 6.47 | 43.6 |
| 5 m | 9.11 | 5.70 | 38.5 |
| 10 m | 9.19 | 5.69 | 38.2 |
| CIgreen | |||
| 0.08 m | 8.87 | 5.95 | 40.1 |
| 0.5 m | 8.24 | 6.63 | 44.6 |
| 1 m | 8.44 | 6.44 | 43.3 |
| 2 m | 8.22 | 6.63 | 44.6 |
| 5 m | 9.12 | 5.69 | 38.4 |
| 10 m | 9.79 | 5.09 | 34.2 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ye, H.; Huang, W.; Huang, S.; Cui, B.; Dong, Y.; Guo, A.; Ren, Y.; Jin, Y. Recognition of Banana Fusarium Wilt Based on UAV Remote Sensing. Remote Sens. 2020, 12, 938. https://doi.org/10.3390/rs12060938
Ye H, Huang W, Huang S, Cui B, Dong Y, Guo A, Ren Y, Jin Y. Recognition of Banana Fusarium Wilt Based on UAV Remote Sensing. Remote Sensing. 2020; 12(6):938. https://doi.org/10.3390/rs12060938
Chicago/Turabian StyleYe, Huichun, Wenjiang Huang, Shanyu Huang, Bei Cui, Yingying Dong, Anting Guo, Yu Ren, and Yu Jin. 2020. "Recognition of Banana Fusarium Wilt Based on UAV Remote Sensing" Remote Sensing 12, no. 6: 938. https://doi.org/10.3390/rs12060938
APA StyleYe, H., Huang, W., Huang, S., Cui, B., Dong, Y., Guo, A., Ren, Y., & Jin, Y. (2020). Recognition of Banana Fusarium Wilt Based on UAV Remote Sensing. Remote Sensing, 12(6), 938. https://doi.org/10.3390/rs12060938