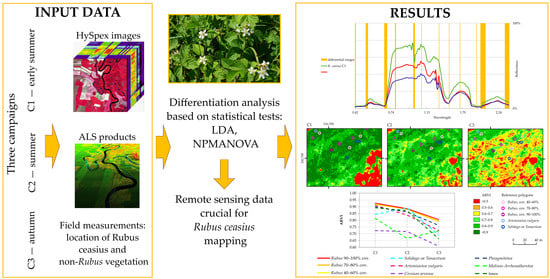
Intra-Annual Variabilities of Rubus caesius L. Discrimination on Hyperspectral and LiDAR Data
Abstract
:1. Introduction
2. Aim of the Study
- What type of remote sensing data differentiates R. caesius from non-Rubus depending on its coverage and the date used for image acquisition;
- Which remote sensing data (spectral bands, calculated indices, and structural metrics derived from ALS) are the most discriminant of R. caesius under different growth and pigmentation phases.
3. Materials and Methods
3.1. Airborne Data Acquisition
3.2. Acquisition and Preprocessing of Ground Reference Measurements
3.3. Airborne Data Processing
3.4. Statistical Analysis
4. Results
4.1. Hyperspectral and ALS Data Comparison
4.2. Spectral Band Dataset
- 416 nm: the first band from blue light, one of the bands with the highest frequency values;
- 515–525 nm: the so-called green edge, where there are absorption bands for carotenoids and partly for chlorophyll;
- 678–707 nm, red-edge: used to determine vegetation conditions;
- 1491 nm: a band with the highest frequency of occurrence as differentiating;
- 1908 nm: water absorption bands;
- 1968–2040 nm and bands above 2300 nm: absorption bands for cellulose and lignin.
4.3. Vegetation Index Dataset
5. Discussion
5.1. Remote Sensing Data that Differentiate Rubus Caesius from Its Background
5.2. Relation between Remote Sensing Data and the Functional Traits of Rubus Caesius
5.3. Remote Sensing Data Useful for Rubus Caesius Identification
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hooper, D.U.; Chapin, F.S.; Ewel, J.J.; Hector, A.; Inchausti, P.; Lavorel, S.; Lawton, J.H.; Lodge, D.M.; Loreau, M.; Naeem, S.; et al. Effects of Biodiversity on Ecosystem Functioning: A Consensus of Current Knowledge. Ecol. Monogr. 2005, 75, 3–35. [Google Scholar] [CrossRef]
- Hejda, M.; Pyšek, P.; Jarošík, V. Impact of Invasive Plants on the Species Richness, Diversity and Composition of Invaded Communities. J. Ecol. 2009, 97, 393–403. [Google Scholar] [CrossRef]
- Van der Putten, W.H. Climate Change, Aboveground-Belowground Interactions, and Species’ Range Shifts. Annu. Rev. Ecol. Evol. Syst. 2012, 43, 365–383. [Google Scholar] [CrossRef]
- Bellard, C.; Thuiller, W.; Leroy, B.; Genovesi, P.; Bakkenes, M.; Courchamp, F. Will Climate Change Promote Future Invasions? Glob. Chang. Biol. 2013, 19, 3740–3748. [Google Scholar] [CrossRef]
- Early, R.; Bradley, B.A.; Dukes, J.S.; Lawler, J.J.; Olden, J.D.; Blumenthal, D.M.; Gonzalez, P.; Grosholz, E.D.; Ibañez, I.; Miller, L.P.; et al. Global Threats from Invasive Alien Species in the Twenty-First Century and National Response Capacities. Nat. Commun. 2016, 7, 12485. [Google Scholar] [CrossRef]
- Marrs, R.H.; Kirby, K.J.; Le Duc, M.G.; McAllister, H.; Smart, S.M.; Oksanen, J.; Bunce, R.G.H.; Corney, P.M. Native Dominants in British Woodland—A Potential Cause of Reduced Species-Richness? New J. Bot. 2013, 3, 156–168. [Google Scholar] [CrossRef] [Green Version]
- Essl, F.; Bacher, S.; Genovesi, P.; Hulme, P.E.; Jeschke, J.M.; Katsanevakis, S.; Kowarik, I.; Kühn, I.; Pyšek, P.; Rabitsch, W.; et al. Which Taxa Are Alien? Criteria, Applications, and Uncertainties. BioScience 2018, 68, 496–509. [Google Scholar] [CrossRef]
- Hejda, M.; Štajerová, K.; Pergl, J.; Pyšek, P. Impacts of Dominant Plant Species on Trait Composition of Communities: Comparison between the Native and Invaded Ranges. Ecosphere 2019, 10. [Google Scholar] [CrossRef]
- Vitousek, P.M. Biological Invasions as Global Environmental Change. Am. Sci. 1996, 84, 468–478. [Google Scholar]
- Williams, D.G.; Baruch, Z. African Grass Invasion in the Americas: Ecosystem Consequences and the Role of Ecophysiology. Biol. Invasions 2000, 2, 123–140. [Google Scholar] [CrossRef]
- Sakai, A.K.; Allendorf, F.W.; Holt, J.S.; Lodge, D.M.; Molofsky, J.; With, K.A.; Baughman, S.; Cabin, R.J.; Cohen, J.E.; Ellstrand, N.C.; et al. The Population Biology of Invasive Species. Annu. Rev. Ecol. Syst. 2001, 32, 305–332. [Google Scholar] [CrossRef] [Green Version]
- Hillebrand, H.; Bennett, D.M.; Cadotte, M.W. Consequences of Dominance: A Review of Evenness Effects on Local and Regional Ecosystem Processes. Ecology 2008, 89, 1510–1520. [Google Scholar] [CrossRef] [PubMed]
- Alice, L.A.; Campbell, C.S. Phylogeny of Rubus (Rosaceae) Based on Nuclear Ribosomal DNA Internal Transcribed Spacer Region Sequences. Am. J. Bot. 1999, 86, 81–97. [Google Scholar] [CrossRef] [PubMed]
- The Plant List. Version 1.1. Available online: http://www.theplantlist.org/ (accessed on 10 May 2020).
- Zielinski, J.; Kosiński, P.; Tomaszewski, D. The Genus Rubus (Rosaceae) in Southeastern Lower Silesia (Poland). Pol. Bot. J. 2004, 49, 161–180. [Google Scholar]
- Oklejewicz, K. Distribution Patterns of Rubus Species (Rosaceae) in the Eastern Part of the Polish Carpathians. Pol. Bot. Stud. 2006, 21, 1–98. [Google Scholar]
- Matuszkiewicz, W. Przewodnik do Oznaczania Zbiorowisk Roślinnych Polski; Wydawnictwo Naukowe PWN: Warsaw, Poland, 2011. [Google Scholar]
- Bâbuţ, C.; Manea, D. Controlling the Perennial Species Rubus Caesius L.—A Problem Weed in Winter Wheat and Grain Maize in Tioişoara Area. Res. J. Agric. Sci. 2010, 42, 8–14. [Google Scholar]
- Bâbuţ, C.; Manea, D. Chemical Control Startegies of Rubus Caesius L. in Grain Maize. Res. J. Agric. Sci. 2010, 42, 3–7. [Google Scholar]
- Edees, E.S.; Newton, A.; Kent, D.H. Brambles of the British Isles; Ray Society: London, UK, 1988. [Google Scholar]
- Wielgosz, T. Wielka Księga Ziół Polskich; Elipsa: Poznań, Poland, 2008. [Google Scholar]
- Dudzinska, D.; Bednarska, K.; Boncler, M.; Luzak, B.; Watala, C. The Influence of Rubus Idaeus and Rubus Caesius Leaf Extracts on Platelet Aggregation in Whole Blood. Cross-Talk of Platelets and Neutrophils. Platelets 2016, 27, 433–439. [Google Scholar] [CrossRef]
- Grochowski, D.M.; Paduch, R.; Wiater, A.; Dudek, A.; Pleszczyńska, M.; Tomczykowa, M.; Granica, S.; Polak, P.; Tomczyk, M. In Vitro Antiproliferative and Antioxidant Effects of Extracts from Rubus Caesius Leaves and Their Quality Evaluation. Evid. Based Complement. Alternat. Med. 2016, 2016, 5698685. [Google Scholar] [CrossRef]
- Widrlechner, M.P.; Wagner, W.H. Occurrence of European Dewberry, Rubus Caesius (Rosaceae), Naturalized in Iowa and Michigan. Mich. Bot. 1998, 37, 107–112. [Google Scholar]
- Mróz, W.; Baba, W. Monitoring of Natural Habitats. Methodological Guide for Natural Habitat 6210 Xerothermic Grasslands (Festuco-Brometea); Library of Environmental Monitoring; Inspection of Environmental Protection: Warsaw, Poland, 2017; pp. 1–14. [Google Scholar]
- Korzeniak, J. Monitoring of Natural Habitats. Methodological Guide for Natural Habitat 6520 Mountain Yellow Trisetum and Bent-Grass Hay Meadows (Polygono Trisetion and Arrhenatherion); Library of Environmental Monitoring; Inspection of Environmental Protection: Warsaw, Poland, 2017; pp. 1–17. [Google Scholar]
- Korzeniak, J. Ekstensywnie Użytkowane Niżowe Łąki Świeże (Arrhenatherion). In Monitoring Siedlisk Przyrodniczych. Przewodnik Metodyczny Część Trzecia; Library of Environmental Monitoring; Inspection of Environmental Protection: Warsaw, Poland, 2012; pp. 79–94. [Google Scholar]
- Pawlaczyk, P. Methodology of Nature Monitoring. Methodological Guide for: Natural Habitats: 4030 Dry Heath Communities Calluno-Genistion, Pohlio-Callunion, Calluno-Arctostaphylion; Library of Environmental Monitoring; Inspection of Environmental Protection: Warsaw, Poland, 2017; pp. 1–19. [Google Scholar]
- Perzanowska, J.; Korzeniak, J.; Chmura, D. Alien Species as a Potential Threat for Natura 2000 Habitats: A National Survey. PeerJ 2019, 7, e8032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dehaan, R.; Louis, J.; Wilson, A.; Hall, A.; Rumbachs, R. Discrimination of Blackberry (Rubus Fruticosus Sp. Agg.) Using Hyperspectral Imagery in Kosciuszko National Park, NSW, Australia. ISPRS J. Photogramm. Remote Sens. 2007, 62, 13–24. [Google Scholar] [CrossRef]
- Sabat-Tomala, A.; Raczko, E.; Zagajewski, B. Comparison of Support Vector Machine and Random Forest Algorithms for Invasive and Expansive Species Classification Using Airborne Hyperspectral Data. Remote Sens. 2020, 12, 516. [Google Scholar] [CrossRef] [Green Version]
- Chance, C.M.; Coops, N.C.; Crosby, K.; Aven, N. Spectral Wavelength Selection and Detection of Two Invasive Plant Species in an Urban Area. Can. J. Remote Sens. 2016, 42, 27–40. [Google Scholar] [CrossRef]
- Chance, C.M.; Coops, N.C.; Plowright, A.A.; Tooke, T.R.; Christen, A.; Aven, N. Invasive Shrub Mapping in an Urban Environment from Hyperspectral and LiDAR-Derived Attributes. Front. Plant Sci. 2016, 07. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verrelst, J.; Geerling, G.W.; Sykora, K.V.; Clevers, J.G.P.W. Mapping of Aggregated Floodplain Plant Communities Using Image Fusion of CASI and LiDAR Data. Int. J. Appl. Earth Obs. Geoinf. 2009, 11, 83–94. [Google Scholar] [CrossRef]
- Rajah, P.; Odindi, J.; Mutanga, O. Evaluating the Potential of Freely Available Multispectral Remotely Sensed Imagery in Mapping American Bramble (Rubus Cuneifolius). S. Afr. Geogr. J. 2018, 100, 291–307. [Google Scholar] [CrossRef]
- Shezi, I.Z.; Poona, N.K. An Investigation into Using Different Satellite Remote Sensors and Techniques to Identify, Map, Monitor and Predict the Spread and Distribution of Some of the Major Current and Emerging Invasive Alien Plant Species in KwaZulu-Natal; Invasive Alien Plant Species Project; UKZN/DEA; School of Environmental Sciences, University of KwaZulu-Natal, Howard College Campus: Durban, South Africa, 2010. [Google Scholar]
- Song, S.; Gong, W.; Zhu, B.; Huang, X. Wavelength Selection and Spectral Discrimination for Paddy Rice, with Laboratory Measurements of Hyperspectral Leaf Reflectance. ISPRS J. Photogramm. Remote Sens. 2011, 66, 672–682. [Google Scholar] [CrossRef]
- Arafat, S.M.; Aboelghar, M.A.; Ahmed, E.F. Crop Discrimination Using Field Hyper Spectral Remotely Sensed Data. Adv. Remote Sens. 2013, 2, 63–70. [Google Scholar] [CrossRef] [Green Version]
- Avola, G.; Di Gennaro, S.F.; Cantini, C.; Riggi, E.; Muratore, F.; Tornambè, C.; Matese, A. Remotely Sensed Vegetation Indices to Discriminate Field-Grown Olive Cultivars. Remote Sens. 2019, 11, 1242. [Google Scholar] [CrossRef] [Green Version]
- Zagajewski, B.; Tømmervik, H.; Bjerke, J.W.; Raczko, E.; Bochenek, Z.; Kłos, A.; Jarocińska, A.; Lavender, S.; Ziółkowski, D. Intraspecific Differences in Spectral Reflectance Curves as Indicators of Reduced Vitality in High-Arctic Plants. Remote Sens. 2017, 9, 1289. [Google Scholar] [CrossRef] [Green Version]
- Sławik, Ł.; Niedzielko, J.; Kania, A.; Piórkowski, H.; Kopeć, D. Multiple Flights or Single Flight Instrument Fusion of Hyperspectral and ALS Data? A Comparison of Their Performance for Vegetation Mapping. Remote Sens. 2019, 11, 970. [Google Scholar] [CrossRef] [Green Version]
- Braun-Blanquet, J. Pflanzensoziologie. Grundzüge der Vegetationskunde, 3rd ed.; Springer-Verlag: Vienna, Austria, New York, NY, USA, 1964. [Google Scholar] [CrossRef]
- RIEGL-RiPROCESS. Available online: http://www.riegl.com/products/software-packages/riprocess/ (accessed on 22 December 2020).
- Terrasolid Software. Available online: https://www.terrasolid.com/ssl/download_software.php (accessed on 12 May 2020).
- HySpex. Available online: https://www.hyspex.com/ (accessed on 18 December 2020).
- PARGE Airborne Image Rectification. Available online: https://www.rese-apps.com/software/parge/index.html (accessed on 18 December 2020).
- Richter, R.; Schlapfer, D. Atmospheric/Topographic Correction for Airborne Imagery; DLR report DLR-IB 565-02/14: Wessling, Germany, 2014. [Google Scholar]
- OrthoVista. Available online: http://www.amigooptima.com/trimble-inpho/ortho-vista.php (accessed on 18 December 2020).
- BCAL Lidar Tools. Available online: https://www.boisestate.edu/bcal/tools-resources/bcal-lidar-tools/ (accessed on 11 May 2020).
- Pfeifer, N.; Mandlburger, G.; Otepka, J.; Karel, W. OPALS—A Framework for Airborne Laser Scanning Data Analysis. Comput. Environ. Urban Syst. 2014, 45, 125–136. [Google Scholar] [CrossRef]
- Bhosale, N.; Manza, R.; Kale, K.V. Analysis of Effect of Gaussian, Salt and Pepper Noise Removal from Noisy Remote Sensing Images. In Second International Conference on Emerging Research in Computing, Information, Communication and Applications (ERCICA 2014); Elsevier: Amsterdam, The Netherlands, 2014. [Google Scholar]
- Shahdoosti, H.R.; Mirzapour, F. Spectral–Spatial Feature Extraction Using Orthogonal Linear Discriminant Analysis for Classification of Hyperspectral Data. Eur. J. Remote Sens. 2017, 50, 111–124. [Google Scholar] [CrossRef]
- R Core Team. R: The R Project for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- Kuhn, M.; Wing, J.; Weston, S.; Williams, A.; Keefer, C.; Engelhardt, A.; Cooper, T.; Mayer, Z.; Kenkel, B.; R Core Team; et al. Caret: Classification and Regression Training; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- Weihs, C.; Ligges, U.; Luebke, K.; Raabe, N. KlaR Analyzing German Business Cycles. In Data Analysis and Decision Support; Baier, D., Decker, R., Schmidt-Thieme, L., Eds.; Springer: Berlin/Heidelberg, Germany, 2005; pp. 335–343. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.; O’Hara, R.; Simpson, G.; Solymos, P.; Stevenes, M.; Wagner, H. Vegan: Community Ecology Package; R Package Version 2.5-6; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- Jensen, J.R. Biophysical Remote Sensing. Ann. Assoc. Am. Geogr. 1983, 73, 111–132. [Google Scholar] [CrossRef]
- Kopeć, D.; Sabat-Tomala, A.; Michalska-Hejduk, D.; Jarocińska, A.; Niedzielko, J. Application of Airborne Hyperspectral Data for Mapping of Invasive Alien Spiraea Tomentosa L.: A Serious Threat to Peat Bog Plant Communities. Wetl. Ecol. Manag. 2020, 28, 357–373. [Google Scholar] [CrossRef] [Green Version]
- Axelsson, A.; Lindberg, E.; Olsson, H. Exploring Multispectral ALS Data for Tree Species Classification. Remote Sens. 2018, 10, 183. [Google Scholar] [CrossRef] [Green Version]
- Ørka, H.O.; Næsset, E.; Bollandsås, O.M. Classifying Species of Individual Trees by Intensity and Structure Features Derived from Airborne Laser Scanner Data. Remote Sens. Environ. 2009, 113, 1163–1174. [Google Scholar] [CrossRef]
- Komárek, J.; Klouček, T.; Prošek, J. The Potential of Unmanned Aerial Systems: A Tool towards Precision Classification of Hard-to-Distinguish Vegetation Types? Int. J. Appl. Earth Obs. Geoinf. 2018, 71, 9–19. [Google Scholar] [CrossRef]
- Prošek, J.; Šímová, P. UAV for Mapping Shrubland Vegetation: Does Fusion of Spectral and Vertical Information Derived from a Single Sensor Increase the Classification Accuracy? Int. J. Appl. Earth Obs. Geoinf. 2019, 75, 151–162. [Google Scholar] [CrossRef]
- De Luca, G.; Silva, J.M.N.; Cerasoli, S.; Araújo, J.; Campos, J.; Di Fazio, S.; Modica, G. Object-Based Land Cover Classification of Cork Oak Woodlands Using UAV Imagery and Orfeo ToolBox. Remote Sens. 2019, 11, 1238. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Wei, Y. Revised Normalized Difference Nitrogen Index (NDNI) for Estimating Canopy Nitrogen Concentration in Wetlands. Optik 2016, 127, 7676–7688. [Google Scholar] [CrossRef]
- Liang, L.; Di, L.; Huang, T.; Wang, J.; Lin, L.; Wang, L.; Yang, M. Estimation of Leaf Nitrogen Content in Wheat Using New Hyperspectral Indices and a Random Forest Regression Algorithm. Remote Sens. 2018, 10, 1940. [Google Scholar] [CrossRef] [Green Version]
- Ellenberg, H.; Weber, H.E.; Düll, R.; Wirth, W.; Werner, W.; Paulißen, D. Zeigerwerte von Pflanzen in Mitteleuropa. Scr. Geobot. 1992, 18, 1–248. [Google Scholar]
- Strik, B.C. Seasonal Variation in Mineral Nutrient Content of Primocane-Fruiting Blackberry Leaves. HortScience 2015, 50, 540–545. [Google Scholar] [CrossRef]
- Hornero, A.; Hernández-Clemente, R.; North, P.R.J.; Beck, P.S.A.; Boscia, D.; Navas-Cortes, J.A.; Zarco-Tejada, P.J. Monitoring the Incidence of Xylella Fastidiosa Infection in Olive Orchards Using Ground-Based Evaluations, Airborne Imaging Spectroscopy and Sentinel-2 Time Series through 3-D Radiative Transfer Modelling. Remote Sens. Environ. 2020, 236, 111480. [Google Scholar] [CrossRef]
- Ponder, A.; Hallmann, E. Phenolics and Carotenoid Contents in the Leaves of Different Organic and Conventional Raspberry (Rubus Idaeus L.) Cultivars and Their In Vitro Activity. Antioxidants 2019, 8, 458. [Google Scholar] [CrossRef] [Green Version]
- Thorp, K.R.; French, A.N.; Rango, A. Effect of Image Spatial and Spectral Characteristics on Mapping Semi-Arid Rangeland Vegetation Using Multiple Endmember Spectral Mixture Analysis (MESMA). Remote Sens. Environ. 2013, 132, 120–130. [Google Scholar] [CrossRef]
- Serbin, G.; Hunt, E.R., Jr.; Daughtry, C.S.T.; McCarty, G.W. Assessment of Spectral Indices for Cover Estimation of Senescent Vegetation. Remote Sens. Lett. 2013, 4, 552–560. [Google Scholar] [CrossRef]
- Wu, W. The Generalized Difference Vegetation Index (GDVI) for Dryland Characterization. Remote Sens. 2014, 6, 1211–1233. [Google Scholar] [CrossRef] [Green Version]
- Solano, F.; Di Fazio, S.; Modica, G. A Methodology Based on GEOBIA and WorldView-3 Imagery to Derive Vegetation Indices at Tree Crown Detail in Olive Orchards. Int. J. Appl. Earth Obs. Geoinf. 2019, 83, 101912. [Google Scholar] [CrossRef]
- Rizeei, H.M.; Shafri, H.Z.M.; Mohamoud, M.A.; Pradhan, B.; Kalantar, B. Oil Palm Counting and Age Estimation from WorldView-3 Imagery and LiDAR Data Using an Integrated OBIA Height Model and Regression Analysis. J. Sens. 2018, 2018, 2536327. Available online: https://www.hindawi.com/journals/js/2018/2536327/ (accessed on 25 December 2020). [CrossRef] [Green Version]
- Varin, M.; Chalghaf, B.; Joanisse, G. Object-Based Approach Using Very High Spatial Resolution 16-Band WorldView-3 and LiDAR Data for Tree Species Classification in a Broadleaf Forest in Quebec, Canada. Remote Sens. 2020, 12, 3092. [Google Scholar] [CrossRef]
- Niphadkar, M.; Nagendra, H. Remote Sensing of Invasive Plants: Incorporating Functional Traits into the Picture. Int. J. Remote Sens. 2016, 37, 3074–3085. [Google Scholar] [CrossRef]









| Field Campaign | Development Phases of Rubus caesius | General Characteristics of the Non-Rubus Vegetation |
|---|---|---|
| C1 | Full development of the above-ground parts of the plant: shoots and leaves, up to approximately 50 cm high; leaves mostly herbaceous and bluish; the vegetative phase predominates; in about 30% of the polygons, R. caesius plants showed the beginning of flowering | Most of the herbaceous vegetation is similar in height to the R. caesius patches; most perennials are in the vegetative stage; some grass species begin flowering |
| C2 | Flowering and fruiting phase; both phases extended over time, not mass-produced flowers or fruits; leaves and shoots herbaceous bluish, glaucous and tinged red where exposed to the sun | Some of the herbaceous plants (Solidago, Phragmites, Tanacetum, Impatiens glandulifera, Artemisia, Urtica) exceed the height of the R. caesius patches (1–1.5 m); plants of some species bloom profusely (Impatiens glandulifera) |
| C3 | Mostly fruiting phase; fruits not mass-produced, did not stand out from the dominant leaves; leaves and steams herbaceous bluish, glaucous and tinged red where exposed to the sun (more intense process than in C2) | Discolouration of leaves of many species (both herbaceous and woody) appear; visible senescence of spring and early summer species plants (drying out) |
| Class of Reference Polygon | C1 (Early Summer) | C2 (Summer) | C3 (Autumn) |
|---|---|---|---|
| R. caesius (coverage 40–60%) | 13 | 1 | 1 |
| R. caesius (coverage 70–80%) | 27 | 23 | 24 |
| R. caesius (coverage 90–100%) | 9 | 22 | 24 |
| Artemisietea vulgaris | 7 | 7 | 7 |
| Cirsium arvense | 9 | 11 | 12 |
| Phragmitetea | 16 | 16 | 16 |
| Molinio-Arrhenatheretea | 8 | 8 | 8 |
| Solidago or Tanacetum | 7 | 8 | 8 |
| Trees (visual interpretation) | 30 | 30 | 30 |
| Sum | 127 | 128 | 131 |
| Scenario | C1 | C2 | C3 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| cov. 40–60% | cov. 70–80% | cov. 90–100% | cov. 70–80% | cov. 90–100% | cov. 70–80% | cov. 90–100% | |||
| Correction rate (based on LDA) | |||||||||
| HS | 0.9651 | 0.9735 | 0.9830 | 0.9491 | 0.9752 | 0.9083 | 0.9321 | ||
| VI | 0.9609 | 0.9816 | 0.9882 | 0.9635 | 0.9808 | 0.9304 | 0.9526 | ||
| OPALS | 0.8571 | 0.8573 | 0.8572 | 0.8577 | 0.8575 | 0.8573 | 0.8572 | ||
| BCAL | 0.8571 | 0.8571 | 0.8572 | 0.8573 | 0.8571 | 0.8577 | 0.8581 | ||
| Number of layers (based on LDA) | |||||||||
| HS | 12 | 11 | 11 | 13 | 17 | 13 | 14 | ||
| VI | 12 | 13 | 12 | 11 | 12 | 11 | 11 | ||
| OPALS | 12 | 12 | 12 | 12 | 13 | 13 | 13 | ||
| BCAL | 10 | 10 | 10 | 11 | 10 | 11 | 11 | ||
| F-value (based on NPMANOVA) | |||||||||
| HS | 9.7 | 10.5 | 12.0 | 8.5 | 9.2 | 4.7 | 6.1 | ||
| C | 18.0 | 16.4 | 23.5 | 23.6 | 42.7 | 9.7 | 11.7 | ||
| OPALS | 1.3 | 1.5 | 1.1 | 1.3 | 1.2 | 1.0 | 1.3 | ||
| BCAL | 1.7 | 1.8 | 1.5 | 1.9 | 2.2 | 1.1 | 1.6 | ||
| VI | Spectrum Used for Calculation in nm | C1 | C2 | C3 | ||||
|---|---|---|---|---|---|---|---|---|
| cov. 40–60% | cov. 70–80% | cov. 90–100% | cov. 70–80% | cov. 90–100% | cov. 70–80% | cov. 90–100% | ||
| ARI1 | 550, 700 | + | + | + | ||||
| ARVI | BLUE, RED, NIR | + | + | + | + | + | ||
| CAI | 1680, 1754 | + | + | + | + | + | + | |
| CRI1 | 510, 550 | + | + | + | + | + | ||
| GDVI | RED, NIR | + | + | |||||
| MRESR | 445, 705 | + | ||||||
| NDNI | 1510, 1680 | + | + | + | + | + | + | + |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jarocińska, A.; Kopeć, D.; Tokarska-Guzik, B.; Raczko, E. Intra-Annual Variabilities of Rubus caesius L. Discrimination on Hyperspectral and LiDAR Data. Remote Sens. 2021, 13, 107. https://doi.org/10.3390/rs13010107
Jarocińska A, Kopeć D, Tokarska-Guzik B, Raczko E. Intra-Annual Variabilities of Rubus caesius L. Discrimination on Hyperspectral and LiDAR Data. Remote Sensing. 2021; 13(1):107. https://doi.org/10.3390/rs13010107
Chicago/Turabian StyleJarocińska, Anna, Dominik Kopeć, Barbara Tokarska-Guzik, and Edwin Raczko. 2021. "Intra-Annual Variabilities of Rubus caesius L. Discrimination on Hyperspectral and LiDAR Data" Remote Sensing 13, no. 1: 107. https://doi.org/10.3390/rs13010107
APA StyleJarocińska, A., Kopeć, D., Tokarska-Guzik, B., & Raczko, E. (2021). Intra-Annual Variabilities of Rubus caesius L. Discrimination on Hyperspectral and LiDAR Data. Remote Sensing, 13(1), 107. https://doi.org/10.3390/rs13010107