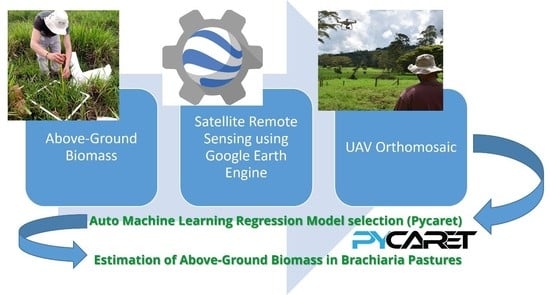
Predictive Modeling of Above-Ground Biomass in Brachiaria Pastures from Satellite and UAV Imagery Using Machine Learning Approaches
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Ground Truth Data (GTD)
2.3. UAV Imagery
2.4. Satellite Imagery
2.5. Multispectral Indices in Remote Sensing
2.6. Satellite and UAV Image Processing
2.7. Modeling and Validation
3. Results
3.1. Data Collection and Feature Extraction
3.2. Machine Learning Model Selection
3.3. Selection of Machine Learning Models Using SATELLITE Data—Description, Analysis, and Tuning
3.4. Selection of Machine Learning Model Using UAV Data—Description, Analysis, and Tuning
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Latham, J.; Cumani, R.; Rosati, I.; Bloise, M. Global Land Cover SHARE Global Land Cover SHARE (GLC-SHARE) Database Beta-Release Version 1.0-2014; 2014. Available online: https://www.fao.org/uploads/media/glc-share-doc.pdf (accessed on 16 October 2022).
- Anderson, J.M. The Effects of Climate Change on Decomposition Processes in Grassland and Coniferous Forests. Ecol. Appl. 1991, 1, 326–347. [Google Scholar] [CrossRef] [PubMed]
- Derner, J.D.; Schuman, G.E. Carbon sequestration and rangelands: A synthesis of land management and precipitation effects. J. Soil Water Conserv. 2007, 62, 77–85. [Google Scholar]
- Habel, J.C.; Dengler, J.; Janišová, M.; Török, P.; Wellstein, C.; Wiezik, M. European grassland ecosystems: Threatened hotspots of biodiversity. Biodivers. Conserv. 2013, 22, 2131–2138. [Google Scholar] [CrossRef] [Green Version]
- Erb, K.-H.; Fetzel, T.; Kastner, T.; Kroisleitner, C.; Lauk, C.; Mayer, A.; Niedertscheider, M.; Erb, K.-H.; Fetzel, T.; Kastner, T.; et al. Livestock Grazing, the Neglected Land Use. Soc. Ecol. 2016, 5, 295–313. [Google Scholar] [CrossRef]
- Fontana, C.S.; Chiarani, E.; da Silva Menezes, L.; Andretti, C.B.; Overbeck, G.E. Bird surveys in grasslands: Do different count methods present distinct results? Rev. Bras. Ornitol. 2018, 26, 116–122. [Google Scholar] [CrossRef]
- Santana, S.S.; Brito, L.F.; Azenha, M.V.; Oliveira, A.A.; Malheiros, E.B.; Ruggieri, A.C.; Reis, R.A. Canopy characteristics and tillering dynamics of Marandu palisade grass pastures in the rainy–dry transition season. Grass Forage Sci. 2017, 72, 261–270. [Google Scholar] [CrossRef]
- Terra, S.; De Andrade Gimenes, F.M.; Giacomini, A.A.; Gerdes, L.; Manço, M.X.; De Mattos, W.T.; Batista, K. Seasonal alteration in sward height of Marandu palisade grass (Brachiaria brizantha) pastures managed by continuous grazing interferes with forage production. Crop Pasture Sci. 2020, 71, 285–293. [Google Scholar] [CrossRef]
- Carnevalli, R.; Silva, S.C.D.; Bueno, A.; Uebele, M.C.; Bueno, F.O.; Hodgson, J.; da Silva, G.N.; Morais, J.G.P.D. Herbage production and grazing losses in Panicum maximum cv. Mombaça under four grazing managements. Trop. Grasslands 2006, 40, 165. [Google Scholar]
- De Oliveira, O.C.; De Oliveira, I.P.; Alves, B.J.R.; Urquiaga, S.; Boddey, R.M. Chemical and biological indicators of decline/degradation of Brachiaria pastures in the Brazilian Cerrado. Agric. Ecosyst. Environ. 2004, 103, 289–300. [Google Scholar] [CrossRef]
- Santos, M.E.R.; da Fonseca, D.M.; Gomes, V.M.; Pimentel, R.M.; Albino, R.L.; da Silva, S.P. Signal grass structure at different sites of the same pasture under three grazing intensities. Acta Sci. Anim. Sci. 2013, 35, 73–78. [Google Scholar] [CrossRef] [Green Version]
- Song, X.-P.; Huang, W.; Hansen, M.C.; Potapov, P. An evaluation of Landsat, Sentinel-2, Sentinel-1 and MODIS data for crop type mapping. Sci. Remote Sens. 2021, 3, 100018. [Google Scholar] [CrossRef]
- Wang, J.; Xiao, X.; Bajgain, R.; Starks, P.; Steiner, J.; Doughty, R.B.; Chang, Q. Estimating leaf area index and aboveground biomass of grazing pastures using Sentinel-1, Sentinel-2 and Landsat images. ISPRS J. Photogramm. Remote Sens. 2019, 154, 189–201. [Google Scholar] [CrossRef]
- Alvarez-Mendoza, C.I.; Teodoro, A.C.; Quintana, J.; Tituana, K. Estimation of Nitrogen in the soil of balsa trees in Ecuador using Unmanned aerial vehicles. In Proceedings of the IEEE IGARSS, Waikoloa, HI, USA, 26 September–2 October 2020; pp. 4610–4613. [Google Scholar]
- De Beurs, K.M.; Henebry, G.M. Land surface phenology, climatic variation, and institutional change: Analyzing agricultural land cover change in Kazakhstan. Remote Sens. Environ. 2004, 89, 497–509. [Google Scholar] [CrossRef]
- Liang, L.; Schwartz, M.D.; Wang, Z.; Gao, F.; Schaaf, C.B.; Tan, B.; Morisette, J.T.; Zhang, X. A cr oss comparison of spatiotemporally enhanced springtime phenological measurements from satellites and ground in a northern U.S. mixed forest. IEEE Trans. Geosci. Remote Sens. 2014, 52, 7513–7526. [Google Scholar] [CrossRef] [Green Version]
- Han, L.; Yang, G.; Dai, H.; Xu, B.; Yang, H.; Feng, H.; Li, Z.; Yang, X. Modeling maize above-ground biomass based on machine learning approaches using UAV remote-sensing data. Plant Methods 2019, 15, 10. [Google Scholar] [CrossRef] [Green Version]
- Filippi, P.; Jones, E.J.; Ginns, B.J.; Whelan, B.M.; Roth, G.W.; Bishop, T.F.A. Mapping the Depth-to-Soil pH Constraint, and the Relationship with Cotton and Grain Yield at the Within-Field Scale. Agronomy 2019, 9, 251. [Google Scholar] [CrossRef] [Green Version]
- Fujisaka, S.; Jones, A. Systems and Farmer Participatory Research: Developments in Research on Natural Resource Management; CIAT Publication: Cali, Colombia, 1999; ISBN 9789586940092. [Google Scholar]
- Benoit, M.; Veysset, P. Livestock unit calculation: A method based on energy needs to refine the study of livestock farming systems. Inra Prod. Anim. 2021, 34, 139–160. [Google Scholar] [CrossRef]
- Alvarez-Mendoza, C.I.; Teodoro, A.; Freitas, A.; Fonseca, J. Spatial estimation of chronic respiratory diseases based on machine learning procedures—An approach using remote sensing data and environmental variables in quito, Ecuador. Appl. Geogr. 2020, 123, 102273. [Google Scholar] [CrossRef]
- Lu, H.; Fan, T.; Ghimire, P.; Deng, L. Experimental Evaluation and Consistency Comparison of UAV Multispectral Minisensors. Remote Sens. 2020, 12, 2542. [Google Scholar] [CrossRef]
- Ventura, D.; Bonifazi, A.; Gravina, M.F.; Belluscio, A.; Ardizzone, G. Mapping and Classification of Ecologically Sensitive Marine Habitats Using Unmanned Aerial Vehicle (UAV) Imagery and Object-Based Image Analysis (OBIA). Remote Sens. 2018, 10, 1331. [Google Scholar] [CrossRef] [Green Version]
- Wulder, M.A.; Loveland, T.R.; Roy, D.P.; Crawford, C.J.; Masek, J.G.; Woodcock, C.E.; Allen, R.G.; Anderson, M.C.; Belward, A.S.; Cohen, W.B.; et al. Current status of Landsat program, science, and applications. Remote Sens. Environ. 2019, 225, 127–147. [Google Scholar] [CrossRef]
- Louis, J.; Pflug, B.; Main-Knorn, M.; Debaecker, V.; Mueller-Wilm, U.; Iannone, R.Q.; Giuseppe Cadau, E.; Boccia, V.; Gascon, F. Sentinel-2 Global Surface Reflectance Level-2a Product Generated with Sen2Cor. Int. Geosci. Remote Sens. Symp. 2019, 8522–8525. [Google Scholar] [CrossRef]
- Panda, S.S.; Ames, D.P.; Panigrahi, S. Application of Vegetation Indices for Agricultural Crop Yield Prediction Using Neural Network Techniques. Remote Sens. 2010, 2, 673–696. [Google Scholar] [CrossRef] [Green Version]
- Xie, Y.; Sha, Z.; Yu, M.; Bai, Y.; Zhang, L. A comparison of two models with Landsat data for estimating above ground grassland biomass in Inner Mongolia, China. Ecol. Modell. 2009, 220, 1810–1818. [Google Scholar] [CrossRef]
- Tuvdendorj, B.; Wu, B.; Zeng, H.; Batdelger, G.; Nanzad, L. Determination of Appropriate Remote Sensing Indices for Spring Wheat Yield Estimation in Mongolia. Remote Sens. 2019, 11, 2568. [Google Scholar] [CrossRef] [Green Version]
- Kenduiywo, B.K.; Carter, M.R.; Ghosh, A.; Hijmans, R.J. Evaluating the quality of remote sensing products for agricultural index insurance. PLoS ONE 2021, 16, e0258215. [Google Scholar] [CrossRef]
- Wu, Q. geemap: A Python package for interactive mapping with Google Earth Engine. J. Open Source Softw. 2020, 5, 2305. [Google Scholar] [CrossRef]
- Selvaraj, M.G.; Valderrama, M.; Guzman, D.; Valencia, M.; Ruiz, H.; Acharjee, A.; Acharjee, A.; Acharjee, A. Machine learning for high-throughput field phenotyping and image processing provides insight into the association of above and below-ground traits in cassava (Manihot esculenta Crantz). Plant Methods 2020, 16, 87. [Google Scholar] [CrossRef]
- Singh, D.; Singh, B. Investigating the impact of data normalization on classification performance. Appl. Soft Comput. 2020, 97, 105524. [Google Scholar] [CrossRef]
- Profillidis, V.A.; Botzoris, G.N. Statistical Methods for Transport Demand Modeling. Model. Transport Demand 2019, 163–224. [Google Scholar] [CrossRef]
- Liu, X. Linear mixed-effects models. Methods Appl. Longitud. Data Anal. 2016, 61–94. [Google Scholar] [CrossRef]
- Moez, A. PyCaret: An open source, low-code machine learning library in Python; 2020. Available online: https://www.pycaret.org (accessed on 16 October 2022).
- Owen, A.B. A robust hybrid of lasso and ridge regression. Contemp. Math. 2007, 443, 59–72. [Google Scholar]
- Mohr, D.L.; Wilson, W.J.; Freund, R.J. Linear Regression. Stat. Methods 2022, 301–349. [Google Scholar] [CrossRef]
- Geurts, P.; Ernst, D.; Wehenkel, L. Extremely randomized trees. Mach. Learn. 2006, 63, 3–42. [Google Scholar] [CrossRef] [Green Version]
- Kalivas, J.H. Data Fusion of Nonoptimized Models: Applications to Outlier Detection, Classification, and Image Library Searching. Data Handl. Sci. Technol. 2019, 31, 345–370. [Google Scholar] [CrossRef]
- Shi, Q.; Abdel-Aty, M.; Lee, J. A Bayesian ridge regression analysis of congestion’s impact on urban expressway safety. Accid. Anal. Prev. 2016, 88, 124–137. [Google Scholar] [CrossRef]
- Kentucky, U. of Rotational vs. Continuous Grazing|Master Grazer. Available online: https://grazer.ca.uky.edu/content/rotational-vs-continuous-grazing (accessed on 3 May 2022).
- Weiss, M.; Jacob, F.; Duveiller, G. Remote sensing for agricultural applications: A meta-review. Remote Sens. Environ. 2020, 236, 111402. [Google Scholar] [CrossRef]
- Pastonchi, L.; Di Gennaro, S.F.; Toscano, P.; Matese, A. Comparison between satellite and ground data with UAV-based information to analyse vineyard spatio-temporal variability. OENO One 2020, 54, 919–934. [Google Scholar] [CrossRef]
- Bretas, I.L.; Valente, D.S.M.; Silva, F.F.; Chizzotti, M.L.; Paulino, M.F.; D’Áurea, A.P.; Paciullo, D.S.C.; Pedreira, B.C.; Chizzotti, F.H.M. Prediction of aboveground biomass and dry-matter content in Brachiaria pastures by combining meteorological data and satellite imagery. Grass Forage Sci. 2021, 76, 340–352. [Google Scholar] [CrossRef]
- Li, B.; Xu, X.; Zhang, L.; Han, J.; Bian, C.; Li, G.; Liu, J.; Jin, L. Above-ground biomass estimation and yield prediction in potato by using UAV-based RGB and hyperspectral imaging. ISPRS J. Photogramm. Remote Sens. 2020, 162, 161–172. [Google Scholar] [CrossRef]
- Marshall, M.; Belgiu, M.; Boschetti, M.; Pepe, M.; Stein, A.; Nelson, A. Field-level crop yield estimation with PRISMA and Sentinel-2. ISPRS J. Photogramm. Remote Sens. 2022, 187, 191–210. [Google Scholar] [CrossRef]
- Abdullah, M.M.; Al-Ali, Z.M.; Srinivasan, S. The use of UAV-based remote sensing to estimate biomass and carbon stock for native desert shrubs. MethodsX 2021, 8, 101399. [Google Scholar] [CrossRef] [PubMed]
- Schucknecht, A.; Seo, B.; Krämer, A.; Asam, S.; Atzberger, C.; Kiese, R. Estimating dry biomass and plant nitrogen concentration in pre-Alpine grasslands with low-cost UAS-borne multispectral data—A comparison of sensors, algorithms, and predictor sets. Biogeosciences 2022, 19, 2699–2727. [Google Scholar] [CrossRef]
- Sharma, P.; Leigh, L.; Chang, J.; Maimaitijiang, M.; Caffé, M. Above-Ground Biomass Estimation in Oats Using UAV Remote Sensing and Machine Learning. Sensors 2022, 22, 601. [Google Scholar] [CrossRef] [PubMed]
- Sabri, Y.; Bahja, F.; Siham, A.; Maizate, A. Cloud Computing in Remote Sensing: Big Data Remote Sensing Knowledge Discovery and Information Analysis. Int. J. Adv. Comput. Sci. Appl. 2021, 12, 888–895. [Google Scholar] [CrossRef]
- Mutanga, O.; Kumar, L. Google Earth Engine Applications. Remote Sens. 2019, 11, 591. [Google Scholar] [CrossRef] [Green Version]
- Torre-Tojal, L.; Bastarrika, A.; Boyano, A.; Lopez-Guede, J.M.; Graña, M. Above-ground biomass estimation from LiDAR data using random forest algorithms. J. Comput. Sci. 2022, 58, 101517. [Google Scholar] [CrossRef]








| Variable | Equipment | Unit |
|---|---|---|
| Height | Flexometer | cm |
| SPAD | SPAD-502Plus | SPAD values |
| Fresh mater (FM) | Gauging with a 0.25 m × 0.25 m frame Precision balance (accuracy: +/− 0.5 g). | gr FM/0.25 m × 0.25 m |
| Dry matter (DM) content production | Precision balance (accuracy: +/− 0.5 g). Sample drying oven | gr DM/0.25 m × 0.25 m |
| Bands | Sentinel-2A | Sentinel-2B | Phantom 4 Multispectral | |||
|---|---|---|---|---|---|---|
| Central Wavelength (nm) | Bandwidth (nm) | Central Wavelength (nm) | Bandwidth (nm) | Central Wavelength (nm) | Bandwidth (nm) | |
| Coastal aerosol | 442.7 | 21 | 442.2 | 21 | - | - |
| Blue (B) | 492.4 | 66 | 492.1 | 66 | 450 | 32 |
| Green (G) | 559.8 | 36 | 559 | 36 | 560 | 32 |
| Red (R) | 664.6 | 31 | 664.9 | 31 | 650 | 32 |
| Vegetation red edge 1 (RE1) | 704.1 | 15 | 703.8 | 16 | - | - |
| Vegetation red edge (RE) | 740.5 | 15 | 739.1 | 15 | 730 | 32 |
| Vegetation red edge 2 (RE2) | 782.8 | 20 | 779.7 | 20 | - | - |
| NIR | 832.8 | 106 | 832.9 | 106 | 840 | 32 |
| Narrow NIR | 864.7 | 21 | 864 | 22 | - | - |
| Water vapor | 945.1 | 20 | 943.2 | 21 | - | - |
| SWIR–Cirrus | 1373.5 | 31 | 1376.9 | 30 | - | - |
| SWIR 1 | 1613.7 | 91 | 1610.4 | 94 | - | - |
| SWIR 2 | 2202.4 | 175 | 2185.7 | 185 | - | - |
| Remote Sensor | Index | Equation–Description |
|---|---|---|
| Satellite-S2 and UAV-P4M | NDRE | (1) |
| NDVI | (2) | |
| GNDVI | (3) | |
| BNDVI | (4) | |
| NPCI | (5) | |
| GRVI | (6) | |
| NGBDI | (7) | |
| P4M | NDREI | (8) |
| CH | Canopy height taken from the DEM by plot | |
| CV | (9) where i, is the pixel associated with the plot | |
| CC_% | Canopy cover is the percent ground cover of the canopy within the pixel surface area |
| Month | Satellite Remote Sensing | GTD | UAV Remote Sensing |
|---|---|---|---|
| July 2021 | 5/7/2021 | 6/7/2021 | |
| 15/7/2021 | 13/7/2021 * | 13/7/2021 | |
| 20/7/2021 | 22/7/2021 | 22/7/2021 | |
| 25/7/2021 | 27/7/2021 * | 27/7/2021 | |
| August 2021 | 4/8/2021 | 6/8/2021 | 6/8/2021 |
| 9/8/2021 | 10/8/2021 * | 10/8/2021 | |
| 19/8/2021 | 17/8/2021 | 17/8/2021 | |
| 24/8/2021 | 24/8/2021 | ||
| 29/8/2021 | 31/8/2021 * | ||
| September 2021 | 8/9/2021 | 7/9/2021 * | 7/9/2021 |
| 13/9/2021 | 15/9/2021 | 15/9/2021 | |
| October 2021 | 3/10/2021 | 5/10/2021 | 5/10/2021 |
| 18/10/2021 | 19/10/2021 | 19/10/2021 | |
| 23/10/2021 | 22/10/2021 * | 22/10/2021 | |
| 28/10/2021 | 26/10/2021–29/10/2021 | 26/10/2021–29/10/2021 | |
| November 2021 | 2/11/2021 | 2/11/2021 | 2/11/2021 |
| 7/11/2021 | 5/11/2021 | ||
| 12/11/2021 | 12/11/2021 * | 12/11/2021 | |
| 17/11/2021 | 16/11/2021 * | 17/11/2021 | |
| 22/11/2021 | 23/11/2021 * | 23/11/2021 | |
| 27/11/2021 | 26/11/2021 * | 26/11/2021 | |
| December 2021 | 2/12/2021 | 3/12/2021 | 3/12/2021 |
| 7/12/2021 | 7/12/2021 | 7/12/2021 | |
| 12/12/2021 | 10/12/2021 * | 10/12/2021 | |
| 22/12/2021 | 23/12/2021 | 23/12/2021 |
| Mean Height | Mean Spad Value | FM | DM | NDVI | NDRE | GNDVI | BNDVI | NPCI | GRVI | NGBDI | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Count | 50 | 50 | 50 | 50 | 50 | 50 | 50 | 50 | 50 | 50 | 50 |
| Mean | 59.92 | 35.54 | 75.3 | 17.94 | 0.69 | 0.47 | 0.60 | 0.72 | 0.03 | 0.19 | 0.40 |
| Std | 12.37 | 2.47 | 51.44 | 13.10 | 0.12 | 0.10 | 0.09 | 0.10 | 0.09 | 0.10 | 0.15 |
| Min | 35.50 | 29.32 | 7 | 1 | 0.39 | 0.26 | 0.39 | 0.49 | −0.18 | 0 | 0.07 |
| 25% | 51.75 | 34.32 | 35.25 | 7 | 0.65 | 0.41 | 0.55 | 0.67 | 0 | 0.10 | 0.35 |
| 50% | 56.75 | 35.24 | 73.50 | 15 | 0.70 | 0.48 | 0.61 | 0.73 | 0.05 | 0.18 | 0.41 |
| 75% | 67.50 | 36.43 | 97.25 | 23.75 | 0.79 | 0.54 | 0.65 | 0.79 | 0.08 | 0.26 | 0.51 |
| MAX | 87.50 | 43.54 | 259 | 57 | 0.89 | 0.69 | 0.77 | 0.86 | 0.31 | 0.37 | 0.69 |
| Height Mean | FM | DM | NDRE_SUM | NDVI_SUM | GNDVI_SUM | BNDVI_SUM | NDREI_SUM | NPCI_SUM | GRVI_SUM | CH_SUM | CV_SUM | CC_% | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Count | 119 | 119 | 119 | 119 | 119 | 119 | 119 | 119 | 119 | 119 | 119 | 119 | 119 |
| Mean | 60.53 | 77.2 | 18.6 | 420,927.42 | 1,466,032.4 | 1,257,027.79 | 1,492,980.23 | 1,270,941.96 | 1,315,352 | 439,031.41 | 206,662,592 | 2,864,893.9 | 71.95 |
| Std | 13.16 | 50.0 | 12.7 | 304,137.5 | 882,413.25 | 805,422.5 | 902,604.36 | 778,299.06 | 800,439.84 | 326,952.07 | 1,006,215,635 | 1,425,754.2 | 30.26 |
| min | 31 | 7 | 1 | −10,497.21 | 38,395.09 | 8998.37 | 48,829.48 | 32,792.05 | 45,411.16 | −46,849.21 | 62,336,892 | 84,975.65 | 2.86 |
| 25% | 52.5 | 34 | 7 | 171,590.09 | 788,830.62 | 606,270.28 | 744,283.75 | 644,213.4 | 643,933.18 | 166,891.06 | 1,376,615,300 | 1,863,291.5 | 54.48 |
| 50% | 58.5 | 79 | 18 | 387,353.7 | 1,378,350.9 | 1,237,617.5 | 1,503,455 | 1,170,034.8 | 1,312,272 | 412,287.44 | 2,075,830,500 | 2,861,639.2 | 81.48 |
| 75% | 67.75 | 103 | 27 | 596,129.93 | 2,035,956.4 | 1,810,215.5 | 2,134,584 | 1,786,781.65 | 1,869,522.8 | 631,205 | 2,765,231,300 | 3,877,652.6 | 97.16 |
| MAX | 94 | 259 | 57 | 1,320,618.1 | 3,201,275.5 | 2,863,656.2 | 3,129,947.2 | 2,819,485.2 | 2,773,591 | 1,179,194.1 | 3,793,030,100 | 5,368,477.5 | 104.0 |
| Dataset | Model | Train R2 | Test R2 | MAE | RMSE |
|---|---|---|---|---|---|
| GTD and Satellite (S2) | Huber Regressor | 0.60 | 0.59 | 0.30 | 0.38 |
| Multiple Linear Regression | 0.54 | 0.63 | 0.34 | 0.43 | |
| Extra Trees Regressor | 0.45 | 0.36 | 0.37 | 0.44 | |
| GTD and UAV (P4M) | K-Nearest Neighbor Regressor | 0.76 | 0.62 | 0.35 | 0.41 |
| Extra Trees Regressor | 0.75 | 0.68 | 0.36 | 0.42 | |
| Bayesian Ridge | 0.70 | 0.61 | 0.37 | 0.45 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alvarez-Mendoza, C.I.; Guzman, D.; Casas, J.; Bastidas, M.; Polanco, J.; Valencia-Ortiz, M.; Montenegro, F.; Arango, J.; Ishitani, M.; Selvaraj, M.G. Predictive Modeling of Above-Ground Biomass in Brachiaria Pastures from Satellite and UAV Imagery Using Machine Learning Approaches. Remote Sens. 2022, 14, 5870. https://doi.org/10.3390/rs14225870
Alvarez-Mendoza CI, Guzman D, Casas J, Bastidas M, Polanco J, Valencia-Ortiz M, Montenegro F, Arango J, Ishitani M, Selvaraj MG. Predictive Modeling of Above-Ground Biomass in Brachiaria Pastures from Satellite and UAV Imagery Using Machine Learning Approaches. Remote Sensing. 2022; 14(22):5870. https://doi.org/10.3390/rs14225870
Chicago/Turabian StyleAlvarez-Mendoza, Cesar I., Diego Guzman, Jorge Casas, Mike Bastidas, Jan Polanco, Milton Valencia-Ortiz, Frank Montenegro, Jacobo Arango, Manabu Ishitani, and Michael Gomez Selvaraj. 2022. "Predictive Modeling of Above-Ground Biomass in Brachiaria Pastures from Satellite and UAV Imagery Using Machine Learning Approaches" Remote Sensing 14, no. 22: 5870. https://doi.org/10.3390/rs14225870
APA StyleAlvarez-Mendoza, C. I., Guzman, D., Casas, J., Bastidas, M., Polanco, J., Valencia-Ortiz, M., Montenegro, F., Arango, J., Ishitani, M., & Selvaraj, M. G. (2022). Predictive Modeling of Above-Ground Biomass in Brachiaria Pastures from Satellite and UAV Imagery Using Machine Learning Approaches. Remote Sensing, 14(22), 5870. https://doi.org/10.3390/rs14225870