Evaluating the Use of Lidar to Discern Snag Characteristics Important for Wildlife
Abstract
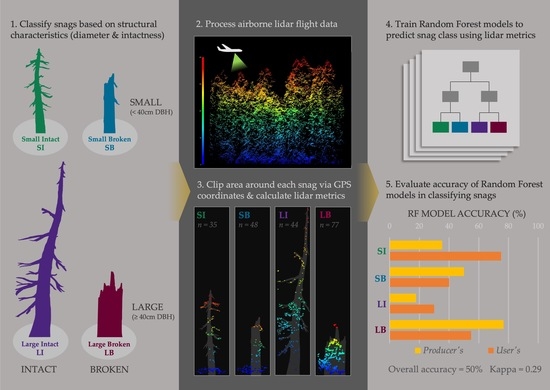
:1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Field Data Collection
2.3. Airborne Lidar Data Acquisition and Processing
2.4. Predictor Variable Selection
2.5. Random Forest Model Development
2.6. Modeling Subset by Diameter
3. Results
3.1. Model Performance across All Four Snag Classes
3.2. Model Performance Subset by Diameter
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Variable | Equation | Ecological significance |
|---|---|---|
| PTDEN | All returns (>1.37 m)]/ total area (19.63 m2) | Relative number of all returns (greater than 1.37 m above ground level, a.g.l.) can reflect the total amount of surface area present to interact with in the canopy, in the form of the snag bole and branches |
| P1stRN | (First returns/all returns) * 100 | Higher percentages of first returns (returns that reflect directly back to sensor) suggest a greater proportion of “external” hits, where lidar has not penetrated the canopy; this may indicate more intact crown features remain |
| P2ndRN | (Second returns/all returns) * 100 | Higher percentages of second returns (returns that bounce once before returning to sensor) suggests a more “porous” snag crown that allows for more pulses to penetrate the canopy |
| P3rdRN | (Third returns/all returns) * 100 | Higher percentages of third returns (returns that bounce twice before returning to sensor) suggests a greater amount of openness mixed with structures within the canopy that allows for pulses to bounce more than once within and still return to the sensor; indicates greater physical complexity |
| GFPmid | Gap Fraction (GF) = Nz/(Ntotal − Nz + dz); N = number of returns, total = across all heights (z), dz = height bin width (1 m); Gap Fraction Profile (GFP) = mean GF across heights; mid = only 5–20m a.g.l. | Gap fraction is calculated as a ratio of the number of points within a given layer versus those that passed through the layer; the relative proportion of canopy gaps within the midcanopy (5–20 m a.g.l.) helps to quantify the total amount of open space present in the lidar footprint at heights where snag crowns will vary, while reducing input from the “gaps” above treetops [40] |
| LADcv | Leaf Area Density (LAD) = −ln(GF)/(k * dz); k = extinction coefficient (0.5), dz = height bin width (2 m); LADcv = LADµ/LADsd | This serves as an indicator of vertical stratification and subdominant structure presence by measuring the homogeneity of vertical strata across all height bins; lower values indicate a more even distribution of vegetation across strata [40] |
| RUMPLE | Surface area of returns (as a convex hull)/ projected area of returns on ground | Higher scores on this index indicate a rougher canopy surface (in terms of heights varying more from one pixel to the next), suggesting greater structural complexity [60] |
| ENTmid | Shannon entropy index (H’) = −∑pz * log(pz); pz = proportion of heights (z), mid = only 5–20m a.g.l. | The evenness of midcanopy heights (5–20 m a.g.l.) may indicate how closed the canopy is; more evenness may suggest an intact dominant canopy, while less evenness may suggest disturbances, such as gaps, subdominant structures such as snags or saplings, or other deviations [61] |
| VAI | Vegetation Area Index (VAI) = ∑LAD | By summing LAD values, this index reports total vegetation coverage in the vertical column; since footprint area is held constant here, this index can be used as a relative measure, where higher scores suggest more canopy, possibly in the form of snag branches and dead needles, or else as encroaching live vegetation (surrounding mature tree crown or sapling) [60] |
| VCI | Vertical Complexity Index (VCI) = (−∑(pz * ln(pz)))/ln(HB); pz = proportion of heights (z), HB = total number of height bins (dz = 1) | Similar to ENTmid (but including all heights), this index measures evenness in terms of proportion of returns by height in the canopy and allows a set maximum height, which can standardize variable snag tops; high scores suggest more even distributions of returns and a higher maximum canopy height, which may mean an intact top [62] |
| ELEV | – | Relative elevation will play a role in forest species composition; as our site was in the Northern Rockies, this can affect tree growth and/or decomposition patterns, as well as affect exposure to higher or lower temperature extremes |
| SLOPE | – | Relative flatness or steepness under a snag may subject it to different disturbance risk levels and types (e.g., steeper slope may have higher winds, leading to more bole break potential) |
| TRASP | – | Relative sun angle interacting with a snag may subject it to different temperature and moisture levels at a microhabitat scale, leading to differing rates of decomposition |
| PTDEN | P1stRN | P2ndRN | P3rdRN | GFPmid | LADcv | RUMPLE | ENTmid | VAI | VCI | ELEV | TRASP | SLOPE | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PTDEN | −0.34 | 0.34 | 0.24 | −0.14 | 0.04 | −0.01 | 0.25 | 0.40 | 0.45 | −0.33 | 0.02 | −0.03 | |
| P1stRN | −0.34 | −0.97 | −0.83 | 0.02 | −0.16 | −0.10 | −0.45 | −0.30 | −0.66 | 0.13 | 0.17 | 0.07 | |
| P2ndRN | 0.34 | −0.97 | 0.68 | −0.05 | 0.15 | 0.11 | 0.39 | 0.32 | 0.62 | −0.21 | −0.13 | −0.09 | |
| P3rdRN | 0.24 | −0.83 | 0.68 | 0.05 | 0.15 | 0.05 | 0.48 | 0.17 | 0.56 | 0.07 | −0.22 | 0.02 | |
| GFPmid | −0.14 | 0.02 | −0.05 | 0.05 | −0.32 | 0.33 | 0.35 | −0.47 | −0.01 | 0.11 | −0.12 | −0.02 | |
| LADcv | 0.04 | −0.16 | 0.15 | 0.15 | −0.32 | −0.42 | 0.07 | 0.20 | 0.21 | 0.07 | 0.00 | 0.06 | |
| RUMPLE | −0.01 | −0.10 | 0.11 | 0.05 | 0.33 | −0.42 | 0.15 | −0.30 | 0.28 | −0.03 | −0.05 | −0.04 | |
| ENTmid | 0.25 | −0.45 | 0.39 | 0.48 | 0.35 | 0.07 | 0.15 | −0.02 | 0.54 | 0.04 | −0.26 | 0.00 | |
| VAI | 0.40 | −0.30 | 0.32 | 0.17 | −0.47 | 0.20 | −0.30 | −0.02 | 0.30 | −0.12 | 0.04 | −0.14 | |
| VCI | 0.45 | −0.66 | 0.62 | 0.56 | −0.01 | 0.21 | 0.28 | 0.54 | 0.30 | −0.13 | −0.18 | −0.07 | |
| ELEV | −0.33 | 0.13 | −0.21 | 0.07 | 0.11 | 0.07 | −0.03 | 0.04 | −0.12 | −0.13 | −0.13 | 0.18 | |
| TRASP | 0.02 | 0.17 | −0.13 | −0.22 | −0.12 | 0.00 | −0.05 | −0.26 | 0.04 | −0.18 | −0.13 | −0.44 | |
| SLOPE | −0.03 | 0.07 | −0.09 | 0.02 | −0.02 | 0.06 | −0.04 | 0.00 | −0.14 | −0.07 | 0.18 | −0.44 |
References
- Tews, J.; Brose, U.; Grimm, V.; Tielbörger, K.; Wichmann, M.C.; Schwager, M.; Jeltsch, F. Animal Species Diversity Driven by Habitat Heterogeneity/Diversity: The Importance of Keystone Structures: Animal Species Diversity Driven by Habitat Heterogeneity. J. Biogeogr. 2004, 31, 79–92. [Google Scholar] [CrossRef] [Green Version]
- Michel, A.K.; Winter, S. Tree Microhabitat Structures as Indicators of Biodiversity in Douglas-Fir Forests of Different Stand Ages and Management Histories in the Pacific Northwest, U.S.A. For. Ecol. Manag. 2009, 257, 1453–1464. [Google Scholar] [CrossRef]
- Larrieu, L.; Paillet, Y.; Winter, S.; Bütler, R.; Kraus, D.; Krumm, F.; Lachat, T.; Michel, A.K.; Regnery, B.; Vandekerkhove, K. Tree Related Microhabitats in Temperate and Mediterranean European Forests: A Hierarchical Typology for Inventory Standardization. Ecol. Indic. 2018, 84, 194–207. [Google Scholar] [CrossRef]
- Jung, K.; Kaiser, S.; Böhm, S.; Nieschulze, J.; Kalko, E.K.V. Moving in Three Dimensions: Effects of Structural Complexity on Occurrence and Activity of Insectivorous Bats in Managed Forest Stands. J. Appl. Ecol. 2012, 49, 523–531. [Google Scholar] [CrossRef]
- Davies, A.B.; Asner, G.P. Advances in Animal Ecology from 3D-LiDAR Ecosystem Mapping. Trends Ecol. Evol. 2014, 29, 681–691. [Google Scholar] [CrossRef]
- Stitt, J.M.; Hudak, A.T.; Silva, C.A.; Vierling, L.A.; Vierling, K.T. Characterizing Individual Tree-level Snags Using Airborne Lidar-derived Forest Canopy Gaps within Closed-canopy Conifer Forests. Methods Ecol. Evol. 2021, 13, 473–484. [Google Scholar] [CrossRef]
- MacArthur, R.H.; MacArthur, J.W. On Bird Species Diversity. Ecology 1961, 42, 594–598. [Google Scholar] [CrossRef]
- Miller, E.; Miller, D.R. Snag Use by Birds; General Technical Report INT-GTR-86; Intermountain Forest and Range Experiment Station, Forest Service, US Department of Agriculture: Ogden, UT, USA, 1980; pp. 337–356. [Google Scholar]
- Sadoti, G.; Pollock, M.G.; Vierling, K.T.; Albright, T.P. Variogram Models Reveal Habitat Gradients Predicting Patterns of Territory Occupancy and Nest Survival among Vesper Sparrows. Wildl. Biol. 2014, 20, 97–107. [Google Scholar] [CrossRef]
- Fielder, P.C.; Starkey, R.G. Bald Eagle Perch-Sites in Eastern Washington. Northwest Sci. 1986, 30, 186–190. [Google Scholar]
- North, M.P.; Franklin, J.F.; Carey, A.B.; Forsman, E.D.; Hamer, T. Forest Stand Structure of the Northern Spotted Owl’s Foraging Habitat. For. Sci. 1999, 45, 520–527. [Google Scholar] [CrossRef]
- Squires, J.R.; Reynolds, R.T. Northern Goshawk (Accipiter gentilis). In The Birds of North America, No. 298; Poole, A., Gill, F., Eds.; The Academy of Natural Sciences Philadelphia, PA: Washington, DC, USA, 1997; pp. 1–31. [Google Scholar]
- Wu, J.X.; Siegel, R.B.; Loffland, H.L.; Tingley, M.W.; Stock, S.L.; Roberts, K.N.; Keane, J.J.; Medley, J.R.; Bridgman, R.; Stermer, C. Diversity of Great Gray Owl Nest Sites and Nesting Habitats in California. J. Wildl. Manag. 2015, 79, 937–947. [Google Scholar] [CrossRef]
- Martinuzzi, S.; Vierling, L.A.; Gould, W.A.; Falkowski, M.J.; Evans, J.S.; Hudak, A.T.; Vierling, K.T. Mapping Snags and Understory Shrubs for a LiDAR-Based Assessment of Wildlife Habitat Suitability. Remote Sens. Environ. 2009, 113, 2533–2546. [Google Scholar] [CrossRef] [Green Version]
- Basile, M.; Asbeck, T.; Pacioni, C.; Mikusiński, G.; Storch, I. Woodpecker Cavity Establishment in Managed Forests: Relative Rather than Absolute Tree Size Matters. Wildl. Biol. 2020, 34, 1453–1466. [Google Scholar] [CrossRef] [Green Version]
- Staniaszek-Kik, M.; Chmura, D.; Żarnowiec, J. What Factors Influence Colonization of Lichens, Liverworts, Mosses and Vascular Plants on Snags? Biologia 2019, 74, 375–384. [Google Scholar] [CrossRef]
- Asbeck, T.; Basile, M.; Stitt, J.; Bauhus, J.; Storch, I.; Vierling, K.T. Tree-Related Microhabitats Are Similar in Mountain Forests of Europe and North America and Their Occurrence May Be Explained by Tree Functional Groups. Trees 2020, 34, 1453–1466. [Google Scholar] [CrossRef]
- Rayner, A.D.M.; Boddy, L. Fungal Decomposition of Wood. Its Biology and Ecology; John Wiley & Sons Ltd.: Chicheste, UK, 1988. [Google Scholar]
- Lorenz, T.J.; Vierling, K.T.; Johnson, T.R.; Fischer, P.C. The Role of Wood Hardness in Limiting Nest Site Selection in Avian Cavity Excavators. Ecol. Appl. 2015, 25, 1016–1033. [Google Scholar] [CrossRef]
- Jusino, M.A.; Lindner, D.L.; Banik, M.T.; Walters, J.R. Heart Rot Hotel: Fungal Communities in Red-Cockaded Woodpecker Excavations. Fungal Ecol. 2015, 14, 33–43. [Google Scholar] [CrossRef]
- Conner, R.N.; Miller, O.K.; Adkisson, C.S. Woodpecker Dependence on Trees Infected by Fungal Heart Rots. Wilson Bull. 1976, 88, 575–581. [Google Scholar]
- Martinuzzi, S.; Vierling, L.A.; Gould, W.A.; Vierling, K.T. Improving the Characterization and Mapping of Wildlife Habitats with Lidar Data: Measurement Priorities for the Inland Northwest, USA. Gap Anal. Bull. 2009, 16, 1–8. [Google Scholar]
- Reitberger, J.; Krzystek, P.; Stilla, U. 3D Segmentation and Classification of Single Trees with Full Waveform LIDAR Data. Proc. SilviLaser 2008, 8, 216–226. [Google Scholar]
- Yao, W.; Krzystek, P.; Heurich, M. Identifying Standing Dead Trees in Forest Areas Based on 3D Single Tree Detection from Full Waveform Lidar Data. ISPRS Ann. Protogrammetry Remote Sens. Spat. Inf. Sci. 2012, 359–364. [Google Scholar] [CrossRef]
- Vauhkonen, J.; Ene, L.; Gupta, S.; Heinzel, J.; Holmgren, J.; Pitkänen, J.; Solberg, S.; Wang, Y.; Weinacker, H.; Hauglin, K.M.; et al. Comparative Testing of Single-Tree Detection Algorithms under Different Types of Forest. For. Int. J. For. Res. 2011, 85, 27–40. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.; Yang, Z.; Cohen, W.B.; Pflugmacher, D.; Lauver, C.L.; Vankat, J.L. Distinguishing between Live and Dead Standing Tree Biomass on the North Rim of Grand Canyon National Park, USA Using Small-Footprint Lidar Data. Remote Sens. Environ. 2009, 113, 2499–2510. [Google Scholar] [CrossRef]
- Bright, B.C.; Hudak, A.T.; McGaughey, R.; Andersen, H.-E.; Negrón, J. Predicting Live and Dead Tree Basal Area of Bark Beetle Affected Forests from Discrete-Return Lidar. Can. J. Remote Sens. 2013, 39, S99–S111. [Google Scholar] [CrossRef]
- Wing, B.M.; Ritchie, M.W.; Boston, K.; Cohen, W.B.; Olsen, M.J. Individual Snag Detection Using Neighborhood Attribute Filtered Airborne Lidar Data. Remote Sens. Environ. 2015, 163, 165–179. [Google Scholar] [CrossRef]
- Casas, Á.; García, M.; Siegel, R.B.; Koltunov, A.; Ramírez, C.; Ustin, S. Burned Forest Characterization at Single-Tree Level with Airborne Laser Scanning for Assessing Wildlife Habitat. Remote Sens. Environ. 2016, 175, 231–241. [Google Scholar] [CrossRef] [Green Version]
- Zellweger, F.; Braunisch, V.; Baltensweiler, A.; Bollmann, K. Remotely Sensed Forest Structural Complexity Predicts Multi Species Occurrence at the Landscape Scale. For. Ecol. Manag. 2013, 307, 303–312. [Google Scholar] [CrossRef]
- Stitt, J.M.; Svancara, L.K.; Vierling, L.A.; Vierling, K.T. Smartphone LIDAR Can Measure Tree Cavity Dimensions for Wildlife Studies. Wildl. Soc. Bull. 2019, 43, 159–166. [Google Scholar] [CrossRef] [Green Version]
- Pesonen, A.; Maltamo, M.; Eerikäinen, K.; Packalèn, P. Airborne Laser Scanning-Based Prediction of Coarse Woody Debris Volumes in a Conservation Area. For. Ecol. Manag. 2008, 255, 3288–3296. [Google Scholar] [CrossRef]
- Krzystek, P.; Serebryanyk, A.; Schnörr, C.; Červenka, J.; Heurich, M. Large-Scale Mapping of Tree Species and Dead Trees in Šumava National Park and Bavarian Forest National Park Using Lidar and Multispectral Imagery. Remote Sens. 2020, 12, 661. [Google Scholar] [CrossRef] [Green Version]
- Bater, C.W.; Coops, N.C.; Gergel, S.E.; Lemay, V.; Collins, D. Estimation of Standing Dead Tree Class Distributions in Northwest Coastal Forests Using Lidar Remote Sensing. Can. J. For. Res. 2009, 39, 1080–1091. [Google Scholar] [CrossRef]
- Idaho Department of Fish and Game (IDFG). Species of Greatest Conservation Need. Available online: https://idfg.idaho.gov/species/taxa/list/sgcn (accessed on 20 September 2021).
- Fekety, P.A.; Falkowski, M.J.; Hudak, A.T.; Jain, T.B.; Evans, J.S. Transferability of Lidar-Derived Basal Area and Stem Density Models within a Northern Idaho Ecoregion. Can. J. Remote Sens. 2018, 44, 131–143. [Google Scholar] [CrossRef]
- Dudley, J.G.; Saab, V. A Field Protocol to Monitor Cavity-Nesting Birds; Res. Pap. RMRS-RP-44; Rocky Mountain Research Station, Forest Service, US Department of Agriculture: Fort Collins, CO, USA, 2003. [Google Scholar]
- Roussel, J.-R.; Auty, D.; Coops, N.C.; Tompalski, P.; Goodbody, T.R.H.; Meador, A.S.; Bourdon, J.-F.; de Boissieu, F.; Achim, A. LidR: An R Package for Analysis of Airborne Laser Scanning (ALS) Data. Remote Sens. Environ. 2020, 251, 112061. [Google Scholar] [CrossRef]
- Vogeler, J.C.; Hudak, A.T.; Vierling, L.A.; Vierling, K.T. Lidar-Derived Canopy Architecture Predicts Brown Creeper Occupancy of Two Western Coniferous Forests. Condor 2013, 115, 614–622. [Google Scholar] [CrossRef]
- Bouvier, M.; Durrieu, S.; Fournier, R.A.; Renaud, J.-P. Generalizing Predictive Models of Forest Inventory Attributes Using an Area-Based Approach with Airborne LiDAR Data. Remote Sens. Environ. 2015, 156, 322–334. [Google Scholar] [CrossRef]
- Liaw, A.; Wiener, M. Classification and Regression by RandomForest. R News 2002, 2, 18–22. [Google Scholar]
- Kuhn, M.; Wing, J.; Weston, S.; Williams, A.; Keefer, C.; Engelhardt, A.; Cooper, T.; Mayer, Z.; Kenkel, B. Others Caret: Classification and Regression Training. R Package Version 6.0-86. Available online: https://cran.r-project.org/web/packages/caret/caret.pdf (accessed on 20 September 2021).
- Davis, J.W. Snags Are for Wildlife; Gen. Tech. Rep. RM-99; Rocky Mountain Forest and Range Experiment Station, Forest Service, US Department of Agriculture: Flagstaff, AZ, USA, 1983; pp. 4–9. [Google Scholar]
- Cohen, J. A Coefficient of Agreement for Nominal Scales. Educ. Psychol. Meas. 1960, 20, 37–46. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Evans, J.S.; Cushman, S.A. Gradient Modeling of Conifer Species Using Random Forests. Landsc. Ecol. 2009, 24, 673–683. [Google Scholar] [CrossRef]
- USGS 3D Elevation Program Topographic Data Quality Levels (QLs). Available online: https://www.usgs.gov/3d-elevation-program/topographic-data-quality-levels-qls (accessed on 20 September 2021).
- Vogeler, J.C.; Hudak, A.T.; Vierling, L.A.; Evans, J.; Green, P.; Vierling, K.T. Terrain and Vegetation Structural Influences on Local Avian Species Richness in Two Mixed-Conifer Forests. Remote Sens. Environ. 2014, 147, 13–22. [Google Scholar] [CrossRef]
- Saab, V.A.; Russell, R.E.; Dudley, J.G. Nest-Site Selection by Cavity-Nesting Birds in Relation to Postfire Salvage Logging. For. Ecol. Manag. 2009, 257, 151–159. [Google Scholar] [CrossRef]
- U.S. Fish & Wildlife Service (US FWS) FWS-Listed U.S. Species by Taxonomic Group—All Animals. Available online: https://ecos.fws.gov/ecp/report/species-listings-by-tax-group?statusCategory=Listed&groupName=All%20Animals (accessed on 20 September 2021).
- Bull, E.L.; Parks, C.G.; Torgersen, T.R. Trees and Logs Important to Wildlife in the Interior Columbia River Basin; Gen. Tech. Rep. PNW-GTR-391; Pacific Northwest Research Station, Forest Service, US Department of Agriculture: Portland, OR, USA, 1997. [Google Scholar]
- McClelland, B.R.; McClelland, P.T. Pileated Woodpecker Nest and Roost Trees in Montana: Links with Old-Growth and Forest “Health”. Wildl. Soc. Bull. 1999, 27, 846–857. [Google Scholar]
- Martin, K.; Aitken, K.E.H.; Wiebe, K.L. Nest Sites and Nest Webs for Cavity-Nesting Communities in Interior British Columbia, Canada: Nest Characteristics and Niche Partitioning. Ornithol. Appl. 2004, 106, 5–19. [Google Scholar] [CrossRef]
- Aitken, K.E.H.; Martin, K. The Importance of Excavators in Hole-Nesting Communities: Availability and Use of Natural Tree Holes in Old Mixed Forests of Western Canada. J. Ornithol. 2007, 148, 425–434. [Google Scholar] [CrossRef]
- Blanc, L.A.; Walters, J.R. Cavity excavation and enlargement as mechanisms for indirect interactions in an avian community. Ecology 2008, 89, 506–514. [Google Scholar] [CrossRef] [Green Version]
- Boucher, P.B.; Hancock, S.; Orwig, D.A.; Duncanson, L.; Armston, J.; Tang, H.; Krause, K.; Cook, B.; Paynter, I.; Li, Z.; et al. Detecting Change in Forest Structure with Simulated GEDI Lidar Waveforms: A Case Study of the Hemlock Woolly Adelgid (HWA; Adelges tsugae) Infestation. Remote Sens. 2020, 12, 1304. [Google Scholar] [CrossRef] [Green Version]
- Hudak, A.T.; Crookston, N.L.; Evans, J.S.; Hall, D.E.; Falkowski, M.J. Nearest Neighbor Imputation of Species-Level, Plot-Scale Forest Structure Attributes from LiDAR Data. Remote Sens. Environ. 2008, 112, 2232–2245. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Lu, C.; Xu, H.; Wang, G. Estimating Aboveground Biomass of Pinus Densata-Dominated Forests Using Landsat Time Series and Permanent Sample Plot Data. J. For. Res. 2019, 30, 1689–1706. [Google Scholar] [CrossRef]
- Neuville, R.; Bates, J.S.; Jonard, F. Estimating Forest Structure from UAV-Mounted LiDAR Point Cloud Using Machine Learning. Remote Sens. 2021, 13, 352. [Google Scholar] [CrossRef]
- LaRue, E.A.; Wagner, F.W.; Fei, S.; Atkins, J.W.; Fahey, R.T.; Gough, C.M.; Hardiman, B.S. Compatibility of Aerial and Terrestrial LiDAR for Quantifying Forest Structural Diversity. Remote Sens. 2020, 12, 1407. [Google Scholar] [CrossRef]
- Pretzsch, H. Forest Dynamics, Growth and Yield; Springer: Berlin/Heidelberg, Germany, 2009; pp. 1–39. [Google Scholar]
- van Ewijk, K.Y.; Treitz, P.M.; Scott, N.A. Characterizing Forest Succession in Central Ontario Using Lidar-Derived Indices. Photogramm. Eng. Remote Sens. 2011, 77, 261–269. [Google Scholar] [CrossRef]



| Parameter | Specification |
|---|---|
| Date collected | 12 October 2016 |
| Vendor | Atlantic Group, LLC |
| Sensor | Leica ALS70-HP |
| Flight altitude | 1965 m above ground level |
| Flight speed | 110 kts |
| Pulse frequency | 278 kHz |
| Scan frequency | 41 Hz |
| Scan angle | ± 30° |
| Swath width | 1098 m |
| Swath overlap | 50% |
| Laser wavelength | 1064 nm |
| Laser beam divergence | 0.22 mrad |
| Vertical accuracy | 5.9 cm |
| Footprint diameter | 43 cm |
| Nominal pulse density | 4.2 pulses/m2 |
| Variable | Description |
|---|---|
| PTDEN | Point density; number of total returns per meter |
| P1stRN | Percent of first returns (out of total returns) |
| P2ndRN | Percent of second returns (out of total returns) |
| P3rdRN | Percent of third returns (out of total returns) |
| GFPmid | Gap Fraction Profile (GFP), calculated only for midcanopy heights (every 1m between 5 m and 20 m above ground) |
| LADcv | Leaf Area Density (LAD) coefficient of variance; derived from measures every 2 m across all canopy heights |
| RUMPLE | Rumple index; roughness of a surface based on ratio between surface area and projected area on the ground |
| ENTmid | Entropy, calculated only for midcanopy heights (between 5 m and 20 m above ground); normalized Shannon diversity and evenness index |
| VAI | Vegetation Area Index (VAI); sum of LAD values, derived from measures every 2 m across all canopy heights |
| VCI | Vertical Complexity Index (VCI); fixed normalization of entropy across heights, derived from measures every 2 m across all canopy heights |
| ELEV | Elevation (in meters), averaged across the 25 m radius reference plot |
| SLOPE | Slope (in degrees), averaged across the 25 m radius reference plot |
| TRASP | Transformed aspect (in degrees), averaged across the 25 m radius reference plot |
| Diameter | Intactness | Number of Snags | Point Density (µ ± sd) |
|---|---|---|---|
| Small (<40 cm) | Intact | 35 | 13.2 ± 7 |
| Broken | 48 | 11.2 ± 5 | |
| Large (≥40 cm) | Intact | 44 | 12.4 ± 5 |
| Broken | 77 | 12.4 ± 7 |
| (a) All Snags | ||||||||
|---|---|---|---|---|---|---|---|---|
| Observed | ||||||||
| Class | SI | SB | LI | LB | Sum | Producer’s Accuracy (%) | User’s Accuracy (%) | |
| Predicted | SI | 6 | 1 | 1 | 0 | 8 | 35.3 | 75.0 |
| SB | 7 | 8 | 3 | 2 | 20 | 50.0 | 40.0 | |
| LI | 1 | 1 | 3 | 5 | 10 | 17.7 | 30.0 | |
| LB | 3 | 6 | 10 | 23 | 42 | 76.7 | 54.8 | |
| Sum | 17 | 16 | 17 | 30 | 80 | |||
| Overall accuracy = 50%; kappa = 0.29 | ||||||||
| Top predictor variables: P3rdRN, LADcv, ELEV | ||||||||
| (b) Small Snags Only | ||||||||
| Observed | ||||||||
| Class | Intact | Broken | Sum | Producer’s Accuracy (%) | User’s Accuracy (%) | |||
| Predicted | Intact | 17 | 3 | 20 | 77.3 | 85.0 | ||
| Broken | 5 | 8 | 13 | 72.7 | 61.5 | |||
| Sum | 22 | 11 | 33 | |||||
| Overall accuracy = 76%; kappa = 0.49 | ||||||||
| Top predictor variables: LADcv, PTDEN, ENTmid | ||||||||
| (c) Large Snags Only | ||||||||
| Observed | ||||||||
| Class | Intact | Broken | Sum | Producer’s Accuracy (%) | User’s Accuracy (%) | |||
| Predicted | Intact | 27 | 9 | 36 | 93.1 | 75.0 | ||
| Broken | 2 | 9 | 11 | 50.0 | 81.8 | |||
| Sum | 29 | 18 | 47 | |||||
| Overall accuracy = 77%; kappa = 0.47 | ||||||||
| Top predictor variables: LADcv, ENTmid, P3rdRN | ||||||||
| (a) All Snags | |||||
|---|---|---|---|---|---|
| Variable | SI | SB | LI | LB | MDGini ↓ |
| P3rdRN | −0.53 | 0.25 | −0.13 | 1.00 | 9.513 |
| LADcv | 0.06 | −0.41 | −0.21 | 0.36 | 8.105 |
| ELEV | 0.46 | 0.43 | 0.07 | −0.11 | 7.636 |
| VAI | 0.01 | −0.21 | 0.51 | −0.07 | 7.445 |
| VCI | −0.25 | −0.04 | −0.08 | 0.47 | 7.365 |
| ENTmid | −0.24 | −0.34 | 0.18 | 0.14 | 7.113 |
| RUMPLE | 0.00 | 0.07 | 0.01 | −0.27 | 6.804 |
| GFPmid | 0.03 | −0.24 | −0.09 | −0.10 | 6.022 |
| SLOPE | 0.28 | 0.30 | 0.00 | 0.09 | 5.964 |
| PTDEN | −0.44 | −0.35 | −0.26 | 0.10 | 5.827 |
| P1stRN | −0.28 | −0.10 | 0.05 | 0.17 | 4.900 |
| P2ndRN | −0.17 | −0.26 | −0.24 | 0.35 | 4.536 |
| TRASP | −0.22 | −0.42 | 0.17 | −0.06 | 4.028 |
| (b) Small Snags Only | |||||
| Variable | Intact | Broken | MDGini ↓ | ||
| LADcv | 0.94 | 0.39 | 3.60 | ||
| PTDEN | 0.98 | 0.64 | 3.12 | ||
| ENTmid | −0.99 | −0.34 | 2.41 | ||
| GFPmid | −0.52 | −1.00 | 2.29 | ||
| ELEV | 0.72 | −0.11 | 1.90 | ||
| SLOPE | −0.14 | −0.51 | 1.81 | ||
| P1stRN | 0.04 | −0.15 | 1.78 | ||
| P3rdRN | −0.42 | −0.04 | 1.57 | ||
| VAI | 0.37 | −0.38 | 1.56 | ||
| TRASP | −0.51 | −0.06 | 1.19 | ||
| VCI | −0.57 | −0.47 | 1.04 | ||
| RUMPLE | −0.97 | −0.44 | 0.94 | ||
| P2ndRN | −0.93 | −0.47 | 0.82 | ||
| (c) Large Snags Only | |||||
| Variable | Intact | Broken | MDGini ↓ | ||
| LADcv | −0.61 | 0.02 | 5.34 | ||
| ENTmid | −0.31 | 0.01 | 3.43 | ||
| P3rdRN | −0.64 | 0.27 | 2.90 | ||
| PTDEN | −0.45 | −0.01 | 2.37 | ||
| ELEV | −0.32 | −0.23 | 2.25 | ||
| RUMPLE | −0.47 | 0.22 | 2.19 | ||
| VCI | −0.69 | −0.10 | 2.15 | ||
| GFPmid | −1.00 | −0.48 | 2.15 | ||
| VAI | −0.72 | −0.38 | 2.01 | ||
| TRASP | −0.35 | −0.15 | 1.97 | ||
| SLOPE | −0.27 | 0.37 | 1.89 | ||
| P2ndRN | −0.55 | 0.18 | 1.47 | ||
| P1stRN | −0.66 | 0.29 | 1.16 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stitt, J.M.; Hudak, A.T.; Silva, C.A.; Vierling, L.A.; Vierling, K.T. Evaluating the Use of Lidar to Discern Snag Characteristics Important for Wildlife. Remote Sens. 2022, 14, 720. https://doi.org/10.3390/rs14030720
Stitt JM, Hudak AT, Silva CA, Vierling LA, Vierling KT. Evaluating the Use of Lidar to Discern Snag Characteristics Important for Wildlife. Remote Sensing. 2022; 14(3):720. https://doi.org/10.3390/rs14030720
Chicago/Turabian StyleStitt, Jessica M., Andrew T. Hudak, Carlos A. Silva, Lee A. Vierling, and Kerri T. Vierling. 2022. "Evaluating the Use of Lidar to Discern Snag Characteristics Important for Wildlife" Remote Sensing 14, no. 3: 720. https://doi.org/10.3390/rs14030720
APA StyleStitt, J. M., Hudak, A. T., Silva, C. A., Vierling, L. A., & Vierling, K. T. (2022). Evaluating the Use of Lidar to Discern Snag Characteristics Important for Wildlife. Remote Sensing, 14(3), 720. https://doi.org/10.3390/rs14030720