A New Approach for Feeding Multispectral Imagery into Convolutional Neural Networks Improved Classification of Seedlings
Abstract
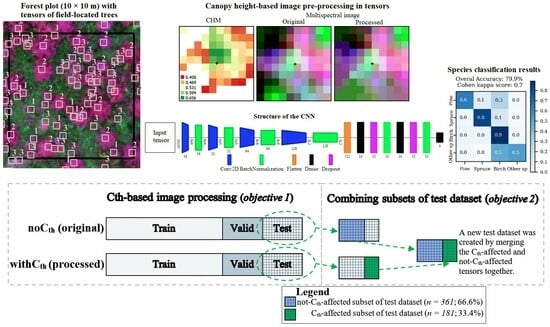
:1. Introduction
- Can the species classification accuracy of seedlings be improved by applying a Cth-based image pre-processing prior to feeding the CNN and RF classifiers?
- Can the species classification accuracy be improved when a new dataset is created by merging the Cth-affected and not-Cth-affected tensors together?
- Can CNN yield a more accurate seedling species classification accuracy than RF due to its higher capability of representing nonlinearity?
- Can species classification be improved by adding vegetation indices (VIs) into multispectral data in CNNs?
2. Materials and Methods
2.1. Study Area and Field Data Collection
2.2. Remote Sensing Data
2.3. Creating Dense Point Clouds and Orthomosaics
2.4. Image Preprocessing
2.5. Extracting Features for the Random Forest Classifier
2.6. Preparing Tensors for the Convolutional Neural Network Classifier
2.7. Training and Validation of the Random Forest Classifier
2.8. Training and Validation of the Convolutional Neural Network
- Dropout rate 1 = [0.0, 0.2, 0.4, 0.6, 0.8];
- Dropout rate 2 = [0.0, 0.2, 0.4, 0.6, 0.8];
- Dense unit 1 = [10, 50, 100, 150, 200, 250, 300];
- Dense unit 2 = [10, 50, 100, 150, 200, 250, 300];
- Batch_size = [32, 64, 128, 256, 1024, 1500].
2.9. Accuracy Assessments
2.10. Analyzing the Effects of the Number of Cth-Affected Cells and Seedlings Height on Classification Accuracy
3. Results
3.1. The Effects of Applying a Canopy Threshold (Cth) on the Accuracy of Species Classification in the CNN and RF Methods
3.2. The Effects of Combining the Subsets of the Test Dataset on the Accuracy of Species Classification in the CNN and RF Methods
3.3. Feature Importance in RF and Configurations of the Classifiers
4. Discussion
4.1. The Effects of Applying Canopy Threshold (Cth)-Based Image Pre-Processing on the Accuracy of Species Classification in CNN and RF
4.2. The Effects of Combining Subsets of Test Dataset on the Accuracy of Species Classification in CNN and RF
4.3. Comparing the Performances of CNN and RF in Seedling–Tree Species Classification
4.4. The Effects of Fusing VIs on the Accuracy of Species Classification in CNN
4.5. The Effects of Seedling-Tree Height and Number of Cth-Affected Pixels on the Accuracy of Species Classification in CNN and RF
4.6. Feature Importance and Model Configurations of the Classifiers
4.7. General Discussion and Future Research
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Layer Type | Output Shape | Number of Parameters |
|---|---|---|
| conv2d_2000 (Conv2D) | (None, 8, 8, 16) | 1888 |
| batch_normalization_3000 | (None, 8, 8, 16) | 64 |
| conv2d_2001 (Conv2D) | (None, 6, 6, 32) | 4640 |
| batch_normalization_3001 | (None, 6, 6, 32) | 128 |
| conv2d_2002 (Conv2D) | (None, 4, 4, 64) | 18,496 |
| batch_normalization_3002 | (None, 4, 4, 64) | 256 |
| conv2d_2003 (Conv2D) | (None, 2, 2, 128) | 73,856 |
| batch_normalization_3003 | (None, 2, 2, 128) | 512 |
| flatten_500 (Flatten) | (None, 512) | 0 |
| dense_1500 (Dense) | (None, 50) | 25,650 |
| dropout_1000 (Dropout) | (None, 50) | 0 |
| batch_normalization_3004 | (None, 50) | 200 |
| dense_1501 (Dense) | (None, 100) | 5100 |
| dropout_1001 (Dropout) | (None, 100) | 0 |
| batch_normalization_3005 | (None, 100) | 400 |
| dense_1502 (Dense) | (None, 4) | 404 |
| Total params: | 131,594 | |
| Trainable params: | 130,814 | |
| Non-trainable params: | 780 |
Appendix B

Appendix C
| Dataset | Classifier | Model Best Param (Out of GridSearch) a | Tunable Param | Total Param | Max Train Accuracy in the Best Model | Max Validation Accuracy in the Best Model | Number of Epochs Ran before Early Stop in the Best Model | Mean Train Time per Epoch in Best Model | St.dev of Train Time per Epoch in Best Model | Total Training Time (GridSearch Time) | Prediction Time |
|---|---|---|---|---|---|---|---|---|---|---|---|
| noCth | RF | None, 0, 2, 2, 1000 | 7776 b | NA | 0.99 | 0.7754 | NA | 36.03 e | 1.94 e | 3 h 32 min | 9 ms |
| CNN noVIs | 32, 0.8, 0.4, 150, 100 | 165,762 | 166,742 | 0.86 | 0.80627 | 300 | 0.63 | 0.07 | 106.8 h (15.25 h) f | 0.3 ms/step | |
| CNN withVIs | 32, 0.6, 0, 100, 50 c | 130,814 | 131,594 | 0.99 | 0.80812 | 133 | 0.75 | 0.19 | 0.3 ms/step | ||
| withCth | RF | None, 0, 1, 2, 1000 | 7776 b | NA | 1.00 | 0.7641 | NA | 8.25 e | 0.38 e | 9 ms | |
| CNN noVIs | 32, 0.6, 0.2, 200, 150 d | 206,862 | 208,042 | 0.99 | 0. 80812 | 119 | 0.64 | 0.11 | 102.8 h (14.68 h) f | 0.3 ms/step | |
| CNN withVIs | 32, 0.8, 0.2, 150, 250 | 266,664 | 267,944 | 0.92 | 0.81550 | 140 | 0.68 | 0.18 | 0.3 ms/step |
References
- Fassnacht, F.E.; Latifi, H.; Stereńczak, K.; Modzelewska, A.; Lefsky, M.; Waser, L.T.; Straub, C.; Ghosh, A. Review of Studies on Tree Species Classification from Remotely Sensed Data. Remote Sens. Environ. 2016, 186, 64–87. [Google Scholar] [CrossRef]
- Imangholiloo, M.; Saarinen, N.; Holopainen, M.; Yu, X.; Hyyppä, J.; Vastaranta, M. Using Leaf-Off and Leaf-On Multispectral Airborne Laser Scanning Data to Characterize Seedling Stands. Remote Sens. 2020, 12, 3328. [Google Scholar] [CrossRef]
- Tapio. Hyvän Metsänhoidon Suositukset. Recommendations for Forest Management in Finland. In Forest Development Centre Tapio; Metsäkustannus Oy: Helsinki, Finland, 2006; 100p. (In Finnish) [Google Scholar]
- Grabska, E.; Socha, J. Evaluating the Effect of Stand Properties and Site Conditions on the Forest Reflectance from Sentinel-2 Time Series. PLoS ONE 2021, 16, e0248459. [Google Scholar] [CrossRef]
- Eriksson, H.M.; Eklundh, L.; Kuusk, A.; Nilson, T. Impact of Understory Vegetation on Forest Canopy Reflectance and Remotely Sensed LAI Estimates. Remote Sens. Environ. 2006, 103, 408–418. [Google Scholar] [CrossRef]
- Joyce, S.; Olsson, H. Monitoring Forest Growth Using Long Time Series of Satellite Data. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. ISPRS Arch. 2000, 33, 1081–1088. [Google Scholar]
- White, J.C.; Coops, N.C.; Wulder, M.A.; Vastaranta, M.; Hilker, T.; Tompalski, P. Remote Sensing Technologies for Enhancing Forest Inventories: A Review. Can. J. Remote Sens. 2016, 42, 619–641. [Google Scholar] [CrossRef]
- Kangas, A.; Astrup, R.; Breidenbach, J.; Fridman, J.; Gobakken, T.; Korhonen, K.T.; Maltamo, M.; Nilsson, M.; Nord-Larsen, T.; Næsset, E.; et al. Remote Sensing and Forest Inventories in Nordic Countries—Roadmap for the Future. Scand. J. For. Res. 2018, 7581, 397–412. [Google Scholar] [CrossRef]
- Hallsby, G.; Ulvcrona, K.A.; Karlsson, A.; Elfving, B.; Sjögren, H.; Ulvcrona, T.; Bergsten, U. Effects of Intensity of Forest Regeneration Measures on Stand Development in a Nationwide Swedish Field Experiment. Forestry 2015, 88, 441–453. [Google Scholar] [CrossRef]
- Korhonen, L.; Pippuri, I.; Packalén, P.; Heikkinen, V.; Maltamo, M.; Heikkilä, J. Detection of the Need for Seedling Stand Tending Using High-Resolution Remote Sensing Data. Silva Fenn. 2013, 47, 105823. [Google Scholar] [CrossRef]
- Uotila, K.; Saksa, T. Effects of Early Cleaning on Young Picea Abies Stands. Scand. J. For. Res. 2014, 29, 111–119. [Google Scholar] [CrossRef]
- Huuskonen, S.; Hynynen, J. Timing and Intensity of Precommercial Thinning and Their Effects on the First Commercial Thinning in Scots Pine Stands. Silva Fenn. 2006, 40, 645–662. [Google Scholar] [CrossRef]
- Kuuluvainen, T.; Gauthier, S. Young and Old Forest in the Boreal: Critical Stages of Ecosystem Dynamics and Management under Global Change. For. Ecosyst. 2018, 5, 26. [Google Scholar] [CrossRef]
- Swanson, M.E.; Franklin, J.F.; Beschta, R.L.; Crisafulli, C.M.; DellaSala, D.A.; Hutto, R.L.; Lindenmayer, D.B.; Swanson, F.J. The Forgotten Stage of Forest Succession: Early-successional Ecosystems on Forest Sites. Front. Ecol. Environ. 2011, 9, 117–125. [Google Scholar] [CrossRef]
- Uotila, K. Optimization of Early Cleaning and Precommercial Thinning Methods in Juvenile Stand Management of Norway Spruce Stands. Ph.D Thesis, Finnish Society of Forest Science, Helsinki, Finland, 2017. [Google Scholar]
- De Lombaerde, E.; Baeten, L.; Verheyen, K.; Perring, M.P.; Ma, S.; Landuyt, D. Understorey Removal Effects on Tree Regeneration in Temperate Forests: A Meta-Analysis. J. Appl. Ecol. 2021, 58, 9–20. [Google Scholar] [CrossRef]
- Dumas, N.; Dupouey, J.L.; Gégout, J.C.; Boulanger, V.; Bontemps, J.D.; Morneau, F.; Dalmasso, M.; Collet, C. Identification and Spatial Extent of Understory Plant Species Requiring Vegetation Control to Ensure Tree Regeneration in French Forests. Ann. For. Sci. 2022, 79, 41. [Google Scholar] [CrossRef]
- Kaila, S.; Kiljunen, N.; Miettinen, A.; Valkonen, S. Effect of Timing of Precommercial Thinning on the Consumption of Working Time in Picea Abies Stands in Finland. Scand. J. For. Res. 2006, 21, 496–504. [Google Scholar] [CrossRef]
- Hynynen, J.; Niemistö, P.; Viherä-Aarnio, A.; Brunner, A.; Hein, S.; Velling, P. Silviculture of Birch (Betula Pendula Roth and Betula Pubescens Ehrh.) in Northern Europe. Forestry 2010, 83, 103–119. [Google Scholar] [CrossRef]
- Martin, M.E.; Newman, S.D.; Aber, J.D.; Congalton, R.G. Determining Forest Species Composition Using High Spectral Resolution Remote Sensing Data. Remote Sens. Environ. 1998, 65, 249–254. [Google Scholar] [CrossRef]
- Maxwell, A.E.; Warner, T.A.; Fang, F. Implementation of Machine-Learning Classification in Remote Sensing: An Applied Review. Int. J. Remote Sens. 2018, 39, 2784–2817. [Google Scholar] [CrossRef]
- Raczko, E.; Zagajewski, B. Comparison of Support Vector Machine, Random Forest and Neural Network Classifiers for Tree Species Classification on Airborne Hyperspectral APEX Images. Eur. J. Remote Sens. 2017, 50, 144–154. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-Learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Ma, L.; Liu, Y.; Zhang, X.; Ye, Y.; Yin, G.; Johnson, B.A. Deep Learning in Remote Sensing Applications: A Meta-Analysis and Review. ISPRS J. Photogramm. Remote Sens. 2019, 152, 166–177. [Google Scholar] [CrossRef]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.W.M.; van Ginneken, B.; Sánchez, C.I. A Survey on Deep Learning in Medical Image Analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef]
- Alzubaidi, L.; Zhang, J.; Humaidi, A.J.; Al-Dujaili, A.; Duan, Y.; Al-Shamma, O.; Santamaría, J.; Fadhel, M.A.; Al-Amidie, M.; Farhan, L. Review of Deep Learning: Concepts, CNN Architectures, Challenges, Applications, Future Directions; Springer International Publishing: Berlin/Heidelberg, Germany, 2021; Volume 8, ISBN 4053702100444. [Google Scholar]
- Gao, Q.; Lim, S.; Jia, X. Hyperspectral Image Classification Using Convolutional Neural Networks and Multiple Feature Learning. Remote Sens. 2018, 10, 299. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, H.; Shen, Q. Spectral-Spatial Classification of Hyperspectral Imagery with 3D Convolutional Neural Network. Remote Sens. 2017, 9, 67. [Google Scholar] [CrossRef]
- Mäyrä, J.; Keski-Saari, S.; Kivinen, S.; Tanhuanpää, T.; Hurskainen, P.; Kullberg, P.; Poikolainen, L.; Viinikka, A.; Tuominen, S.; Kumpula, T.; et al. Tree Species Classification from Airborne Hyperspectral and LiDAR Data Using 3D Convolutional Neural Networks. Remote Sens. Environ. 2021, 256, 112322. [Google Scholar] [CrossRef]
- Natesan, S.; Armenakis, C.; Vepakomma, U. Individual Tree Species Identification Using Dense Convolutional Network (Densenet) on Multitemporal RGB Images from UAV. J. Unmanned Veh. Syst. 2020, 8, 310–333. [Google Scholar] [CrossRef]
- Nezami, S.; Khoramshahi, E.; Nevalainen, O.; Pölönen, I.; Honkavaara, E. Tree Species Classification of Drone Hyperspectral and RGB Imagery with Deep Learning Convolutional Neural Networks. Remote Sens. 2020, 12, 1070. [Google Scholar] [CrossRef]
- Yan, S.; Jing, L.; Wang, H. A New Individual Tree Species Recognition Method Based on a Convolutional Neural Network and High-spatial Resolution Remote Sensing Imagery. Remote Sens. 2021, 13, 479. [Google Scholar] [CrossRef]
- Fricker, G.A.; Ventura, J.D.; Wolf, J.A.; North, M.P.; Davis, F.W.; Franklin, J. A Convolutional Neural Network Classifier Identifies Tree Species in Mixed-Conifer Forest from Hyperspectral Imagery. Remote Sens. 2019, 11, 2326. [Google Scholar] [CrossRef]
- Pleşoianu, A.I.; Stupariu, M.S.; Şandric, I.; Pătru-Stupariu, I.; Drăguţ, L. Individual Tree-Crown Detection and Species Classification in Very High-Resolution Remote Sensing Imagery Using a Deep Learning Ensemble Model. Remote Sens. 2020, 12, 2426. [Google Scholar] [CrossRef]
- Guo, X.; Li, H.; Jing, L.; Wang, P. Individual Tree Species Classification Based on Convolutional Neural Networks and Multitemporal High-Resolution Remote Sensing Images. Sensors 2022, 22, 3157. [Google Scholar] [CrossRef]
- Li, H.; Hu, B.; Li, Q.; Jing, L. Cnn-Based Individual Tree Species Classification Using High-Resolution Satellite Imagery and Airborne Lidar Data. Forests 2021, 12, 1697. [Google Scholar] [CrossRef]
- Pearse, G.D.; Tan, A.Y.S.; Watt, M.S.; Franz, M.O.; Dash, J.P. Detecting and Mapping Tree Seedlings in UAV Imagery Using Convolutional Neural Networks and Field-Verified Data. ISPRS J. Photogramm. Remote Sens. 2020, 168, 156–169. [Google Scholar] [CrossRef]
- Hartley, R.J.L.; Leonardo, E.M.; Massam, P.; Watt, M.S.; Estarija, H.J.; Wright, L.; Melia, N.; Pearse, G.D. An Assessment of High-Density UAV Point Clouds for the Measurement of Young Forestry Trials. Remote Sens. 2020, 12, 4039. [Google Scholar] [CrossRef]
- Chadwick, A.J.; Goodbody, T.R.H.; Coops, N.C.; Hervieux, A.; Bater, C.W.; Martens, L.A.; White, B.; Röeser, D. Automatic Delineation and Height Measurement of Regenerating Conifer Crowns under Leaf-off Conditions Using Uav Imagery. Remote Sens. 2020, 12, 4104. [Google Scholar] [CrossRef]
- Fromm, M.; Schubert, M.; Castilla, G.; Linke, J.; McDermid, G. Automated Detection of Conifer Seedlings in Drone Imagery Using Convolutional Neural Networks. Remote Sens. 2019, 11, 2585. [Google Scholar] [CrossRef]
- Næsset, E.; Bjerknes, K.-O. Estimating Tree Heights and Number of Stems in Young Forest Stands Using Airborne Laser Scanner Data. Remote Sens. Environ. 2001, 78, 328–340. [Google Scholar] [CrossRef]
- Korpela, I.; Tuomola, T.; Tokola, T.; Dahlin, B. Appraisal of Seedling Stand Vegetation with Airborne Imagery and Discrete-Return LiDAR—an Exploratory Analysis. Silva Fenn. 2008, 42, 753–772. [Google Scholar] [CrossRef]
- Økseter, R.; Bollandsås, O.M.; Gobakken, T.; Næsset, E. Modeling and Predicting Aboveground Biomass Change in Young Forest Using Multi-Temporal Airborne Laser Scanner Data. Scand. J. For. Res. 2015, 30, 458–469. [Google Scholar] [CrossRef]
- Imangholiloo, M.; Yrttimaa, T.; Mattsson, T.; Junttila, S.; Holopainen, M.; Saarinen, N.; Savolainen, P.; Hyyppä, J.; Vastaranta, M. Adding Single Tree Features and Correcting Edge Tree Effects Enhance the Characterization of Seedling Stands with Single-Photon Airborne Laser Scanning. ISPRS J. Photogramm. Remote Sens. 2022, 191, 129–142. [Google Scholar] [CrossRef]
- Imangholiloo, M.; Saarinen, N.; Markelin, L.; Rosnell, T.; Näsi, R.; Hakala, T.; Honkavaara, E.; Holopainen, M.; Hyyppä, J.; Vastaranta, M. Characterizing Seedling Stands Using Leaf-Off and Leaf-On Photogrammetric Point Clouds and Hyperspectral Imagery Acquired from Unmanned Aerial Vehicle. Forests 2019, 10, 415. [Google Scholar] [CrossRef]
- Falbel, D.; Allaire, J.J.; Chollet, F. Keras Open-Source Neural-Network Library Written in Python v2.13.1. 2015. Available online: https://github.com/keras-team/keras (accessed on 31 January 2023).
- Abadi, M.; Barham, P.; Chen, J.; Chen, Z.; Davis, A.; Dean, J.; Devin, M.; Ghemawat, S.; Irving, G.; Isard, M.; et al. Tensorflow: A System for Large-Scale Machine Learning. In Proceedings of the 12th Symposium on Operating Systems Design and ImplementationSymposium on Operating Systems Design and Implementation, Savannah, GA, USA, 2–4 November 2016; pp. 265–283. [Google Scholar]
- Bauerle, A.; Van Onzenoodt, C.; Ropinski, T. Net2Vis-A Visual Grammar for Automatically Generating Publication-Tailored CNN Architecture Visualizations. IEEE Trans. Vis. Comput. Graph. 2021, 27, 2980–2991. [Google Scholar] [CrossRef] [PubMed]
- Hinton, G.E.; Srivastava, N.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R.R. Improving Neural Networks by Preventing Co-Adaptation of Feature Detectors. arXiv 2012, arXiv:1207.0580. [Google Scholar]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A Simple Way to Prevent Neural Networks from Overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Varshney, M.; Singh, P. Optimizing Nonlinear Activation Function for Convolutional Neural Networks. Signal, Image Video Process. 2021, 15, 1323–1330. [Google Scholar] [CrossRef]
- CSC—IT Center for Science Finland. Supercomputer Puhti Is Now Available for Researchers—Supercomputer Puhti Is Now Available for Researchers. CSC Company Site. 2022. Available online: https://www.csc.fi/En/-/Supertietokone-Puhti-on-Avattu-Tutkijoiden-Kayttoon (accessed on 31 January 2023).
- Prasad, M.P.S.; Senthilrajan, A. A Novel CNN-KNN Based Hybrid Method for Plant Classification. J. Algebr. Stat. 2022, 13, 498–502. [Google Scholar]
- Chen, C.; Jing, L.; Li, H.; Tang, Y. A New Individual Tree Species Classification Method Based on the Resu-Net Model. Forests 2021, 12, 1202. [Google Scholar] [CrossRef]
- Zhang, B.; Zhao, L.; Zhang, X. Three-Dimensional Convolutional Neural Network Model for Tree Species Classification Using Airborne Hyperspectral Images. Remote Sens. Environ. 2020, 247, 111938. [Google Scholar] [CrossRef]
- Ao, L.; Feng, K.; Sheng, K.; Zhao, H.; He, X.; Chen, Z. TPENAS: A Two-Phase Evolutionary Neural Architecture Search for Remote Sensing Image Classification. Remote Sens. 2023, 15, 2212. [Google Scholar] [CrossRef]
- Martins, G.B.; La Rosa, L.E.C.; Happ, P.N.; Filho, L.C.T.C.; Santos, C.J.F.; Feitosa, R.Q.; Ferreira, M.P. Deep Learning-Based Tree Species Mapping in a Highly Diverse Tropical Urban Setting. Urban For. Urban Green. 2021, 64, 127241. [Google Scholar] [CrossRef]
- Anderson, C.J.; Heins, D.; Pelletier, K.C.; Knight, J.F. Improving Machine Learning Classifications of Phragmites Australis Using Object-Based Image Analysis. Remote Sens. 2023, 15, 989. [Google Scholar] [CrossRef]
- Xi, Y.; Ren, C.; Wang, Z.; Wei, S.; Bai, J.; Zhang, B.; Xiang, H.; Chen, L. Mapping Tree Species Composition Using OHS-1 Hyperspectral Data and Deep Learning Algorithms in Changbai Mountains, Northeast China. Forests 2019, 10, 818. [Google Scholar] [CrossRef]
- Hartling, S.; Sagan, V.; Sidike, P.; Maimaitijiang, M.; Carron, J. Urban Tree Species Classification Using a Worldview-2/3 and LiDAR Data Fusion Approach and Deep Learning. Sensors 2019, 19, 1284. [Google Scholar] [CrossRef]
- Ye, N.; Morgenroth, J.; Xu, C.; Chen, N. Indigenous Forest Classification in New Zealand—A Comparison of Classifiers and Sensors. Int. J. Appl. Earth Obs. Geoinf. 2021, 102, 102395. [Google Scholar] [CrossRef]
- Adagbasa, E.G.; Adelabu, S.A.; Okello, T.W. Application of Deep Learning with Stratified K-Fold for Vegetation Species Discrimation in a Protected Mountainous Region Using Sentinel-2 Image. Geocarto Int. 2022, 37, 142–162. [Google Scholar] [CrossRef]
- Trier, Ø.D.; Salberg, A.B.; Kermit, M.; Rudjord, Ø.; Gobakken, T.; Næsset, E.; Aarsten, D. Tree Species Classification in Norway from Airborne Hyperspectral and Airborne Laser Scanning Data. Eur. J. Remote Sens. 2018, 51, 336–351. [Google Scholar] [CrossRef]
- Yaloveha, V.; Hlavcheva, D.; Podorozhniak, A. Spectral Indexes Evaluation for Satellite Images Classification Using CNN. J. Inf. Organ. Sci. 2021, 45, 435–449. [Google Scholar] [CrossRef]
- Zhu, Y.; Liu, K.; Liu, L.; Myint, S.W.; Wang, S.; Liu, H.; He, Z. Exploring the Potential of World View-2 Red-Edge Band-Based Vegetation Indices for Estimation of Mangrove Leaf Area Index with Machine Learning Algorithms. Remote Sens. 2017, 9, 1060. [Google Scholar] [CrossRef]
- Sun, Y.; Xin, Q.; Huang, J.; Huang, B.; Zhang, H. Characterizing Tree Species of a Tropical Wetland in Southern China at the Individual Tree Level Based on Convolutional Neural Network. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2019, 12, 4415–4425. [Google Scholar] [CrossRef]












| Multispectral Images | RGB Images | |
|---|---|---|
| Name of Sensor | MicaSense MX Red-Edge | Sony A6000-series 24 Megapixel frame camera with 21 mm Voigtländer lens |
| Spectral bands | RGB, RedEdge, NIR | RGB |
| Central wavelength (nm) | 475, 560, 668, 717, 840 | - |
| FWHM (nm) | 20, 20, 10, 10, 40 | - |
| Resolution (GSD, cm) | 5.5 | 1.3 |
| Flight Altitude (m) | 70 | 70 |
| Drone speed (m/s) | 8–9 | 8–9 |
| Date | 11 * and 15 ** September 2021 | 11 * and 15 ** September 2021 |
| Time (UTC + 3) | 12:30 to 14:30 * and 10:30 to 11:50 ** | 12:30 to 14:30 * and 10:30 to 11:50 ** |
| Forward and side overlap (%) | 80 and 75 | 85 and 80 |
| Feature Name | Description of Features Meaning |
|---|---|
| Min | Minimum of reflectance within pixels of each tensor. |
| Max | Maximum of reflectance within pixels of each tensor. |
| Mean | Mean of reflectance within pixels of each tensor. |
| Stdev | Standard deviation of reflectance within pixels of each tensor. |
| Range | Range (Max-Min) of reflectance within pixels of each tensor. |
| Percentiles | Percentiles of the reflectance values of pixels of each tensor. Percentiles 10–90 (every 10%) and percentiles 5 and 95% were calculated, totaling 11 features. |
| NoData | Number of NoData pixels of each tensor which were omitted assuming as understory reflectance. |
| Full Name | Abbreviation | Equation | |
|---|---|---|---|
| 1 | Normalized Difference Vegetation Index | NDVI | (NIR − Red)/(NIR + Red) |
| 2 | RedEdge NDVI | NDRE | (NIR − RedEdge)/(NIR + RedEdge) |
| 3 | Green NDVI | GNDVI | (NIR − Green)/(NIR + Green) |
| 4 | Simple ratio | SR | NIR/Red |
| 5 | NDVI times SR | NDVI × SR | NDVI × SR |
| 6 | Chlorophyll Vegetation Index | CVI | (NIR/Green) × (Red/Green) |
| 7 | Normalized Difference Greenness Index | NDGI | Green − Red/Green + Red |
| 8 | Difference Vegetation Index | DVI | NIR − Red |
| Species | Number (%) of Training Set | Number (%) of Validation Set | Number (%) of Test Set |
|---|---|---|---|
| Pine | 579 (13.4%) | 79 (14.6%) | 81 (14.9%) |
| Spruce | 1255 (29.0%) | 150 (27.7%) | 149 (27.5%) |
| Birch | 2103 (48.5%) | 258 (47.6%) | 261 (48.2%) |
| Other species | 396 (9.1%) | 55 (10.1%) | 51 (9.4%) |
| Total (5417) | 4333 (80.0%) | 542 (10.0%) | 542 (10.0%) |
| Parameter Name | Description (Pedregosa et al. [24]) | Given Values for Grid | Default Value |
|---|---|---|---|
| max_depth | The maximum depth of the tree. | [None, 2, 10, 50, 80, 100] | None |
| min_samples_split | The minimum number of samples required to split an internal node. | [2, 3, 5, 8, 10, 12] | 2 * |
| min_samples_leaf | The minimum number of samples required to be at a leaf node. | [1, 2, 3, 5, 20, 100] | 1 |
| max_features | The number of features to consider when looking for the best split ** | [0, 2, ‘auto’, ‘log2’, ‘sqrt’, None] | sqrt |
| n_estimators | The number of trees in the forest. Usually, the bigger the better, but a larger number slows down the computation. | [75, 100, 125, 200, 500, 1000] | 100 |
| Dataset | Classifier | OA (%) | Kappa | Overall Precision (Per Species) * | Overall Recall (Per Species) * | Overall F1 Macro (Per Species) * | Overall F1 Micro |
|---|---|---|---|---|---|---|---|
| NoCth | RF | 67.9 | 0.5 | 0.7 (0.5, 0. 7, 0.7, 1.0) | 0.6 (0.3, 0.7, 0.8, 0.4) | 0.6 (0.3, 0.7, 0.8, 0.6) | 0.7 |
| CNN noVIs | 76.9 | 0.6 | 0.8 (0.7, 0.8, 0.8, 0.8) | 0.7 (0.6, 0.8, 0.9, 0.4) | 0.7 (0.6, 0.8, 0.8, 0.5) | 0.8 | |
| CNN withVIs | 79.0 | 0.7 | 0.8 (0.7, 0.9, 0. 8, 0.7) | 0.7 (0.6, 0.8, 0.9, 0.5) | 0.7 (0.7, 0.9, 0.8, 0.6) | 0.8 | |
| WithCth | RF | 68.3 | 0.5 | 0.7 (0.6, 0.7, 0.7, 0.8) | 0.7 (0.3, 0.8, 0.8, 0.4) | 0.6 (0.4, 0.7, 0.8, 0.5) | 0.7 |
| CNN noVIs | 75.1 | 0.6 | 0.7 (0.7, 0.8, 0.7, 0.7) | 0.7 (0.6, 0.8, 0.8, 0.4) | 0.7 (0.6, 0.8, 0.8, 0.5) | 0.8 | |
| CNN withVIs | 79.3 | 0.7 | 0.8 (0.8, 0.8, 0.8, 0.8) | 0.7 (0.6, 0.8, 0.9, 0.5) | 0.7 (0.7, 0.8, 0.8, 0.6) | 0.8 | |
| Combined dataset | RF | 66.6 | 0.5 | 0.7 (0.6, 0.6, 0.7, 0.9) | 0.5 (0.2, 0.7, 0.9, 0.4) | 0.6 (0.3, 0.7, 0.8, 0.6) | 0.7 |
| CNN noVIs | 77.3 | 0.6 | 0.8 (0.7, 0.8, 0.8, 0.8) | 0.7 (0.5, 0.8, 0.9, 0.4) | 0.7 (0.6, 0.8, 0.8, 0.5) | 0.8 | |
| CNN withVIs | 79.9 | 0.7 | 0.8 (0.8, 0.9, 0. 8, 0.7) | 0.7 (0.6, 0.8, 0.9, 0.5) | 0.7 (0.7, 0.9, 0.8, 0.6) | 0.8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Imangholiloo, M.; Luoma, V.; Holopainen, M.; Vastaranta, M.; Mäkeläinen, A.; Koivumäki, N.; Honkavaara, E.; Khoramshahi, E. A New Approach for Feeding Multispectral Imagery into Convolutional Neural Networks Improved Classification of Seedlings. Remote Sens. 2023, 15, 5233. https://doi.org/10.3390/rs15215233
Imangholiloo M, Luoma V, Holopainen M, Vastaranta M, Mäkeläinen A, Koivumäki N, Honkavaara E, Khoramshahi E. A New Approach for Feeding Multispectral Imagery into Convolutional Neural Networks Improved Classification of Seedlings. Remote Sensing. 2023; 15(21):5233. https://doi.org/10.3390/rs15215233
Chicago/Turabian StyleImangholiloo, Mohammad, Ville Luoma, Markus Holopainen, Mikko Vastaranta, Antti Mäkeläinen, Niko Koivumäki, Eija Honkavaara, and Ehsan Khoramshahi. 2023. "A New Approach for Feeding Multispectral Imagery into Convolutional Neural Networks Improved Classification of Seedlings" Remote Sensing 15, no. 21: 5233. https://doi.org/10.3390/rs15215233
APA StyleImangholiloo, M., Luoma, V., Holopainen, M., Vastaranta, M., Mäkeläinen, A., Koivumäki, N., Honkavaara, E., & Khoramshahi, E. (2023). A New Approach for Feeding Multispectral Imagery into Convolutional Neural Networks Improved Classification of Seedlings. Remote Sensing, 15(21), 5233. https://doi.org/10.3390/rs15215233